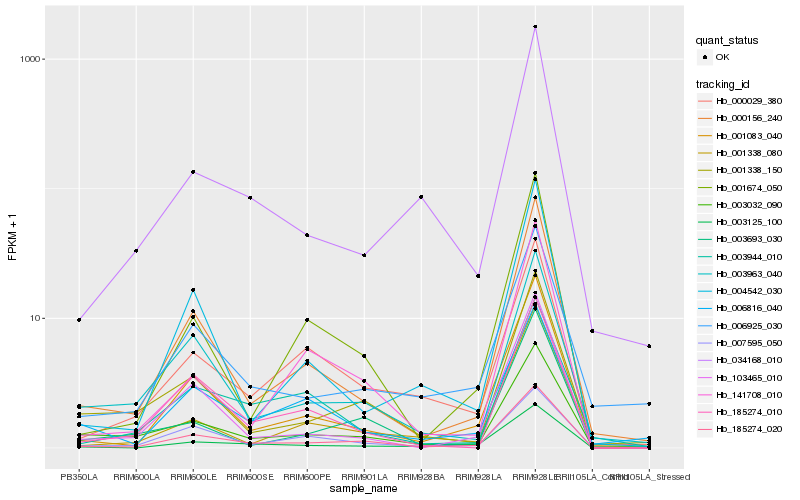
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185274_020 |
0.0 |
- |
- |
Ycf2 (chloroplast) [Andrographis paniculata] |
| 2 |
Hb_001083_040 |
0.1069490633 |
- |
- |
hypothetical protein EUGRSUZ_E00717 [Eucalyptus grandis] |
| 3 |
Hb_003125_100 |
0.1192549327 |
- |
- |
NAD(P)-binding Rossmann-fold superfamily protein [Theobroma cacao] |
| 4 |
Hb_000156_240 |
0.1311428568 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 5 |
Hb_003032_090 |
0.1352939382 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 6 |
Hb_000029_380 |
0.1359892178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_185274_010 |
0.145255285 |
- |
- |
hypothetical protein ARALYDRAFT_330174 [Arabidopsis lyrata subsp. lyrata] |
| 8 |
Hb_007595_050 |
0.1453591999 |
- |
- |
hypothetical chloroplast RF2 [Hevea brasiliensis] |
| 9 |
Hb_103465_010 |
0.1467769742 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 10 |
Hb_003963_040 |
0.1474794725 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 11 |
Hb_001338_150 |
0.1484964179 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 12 |
Hb_001674_050 |
0.1509697899 |
- |
- |
cytochrome b6 (chloroplast) [Apios americana] |
| 13 |
Hb_006925_030 |
0.1571529178 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 14 |
Hb_001338_080 |
0.1588565764 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Tamarindus indica] |
| 15 |
Hb_006816_040 |
0.1589264191 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 16 |
Hb_003693_030 |
0.1617347564 |
- |
- |
NADH dehydrogenase subunit 7 [Hevea brasiliensis] |
| 17 |
Hb_034168_010 |
0.162415729 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 18 |
Hb_141708_010 |
0.1663787458 |
- |
- |
hypothetical protein EUTSA_v10011731mg [Eutrema salsugineum] |
| 19 |
Hb_004542_030 |
0.1681748882 |
- |
- |
hypothetical protein AMTR_s00334p00014280 [Amborella trichopoda] |
| 20 |
Hb_003944_010 |
0.1708022691 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |