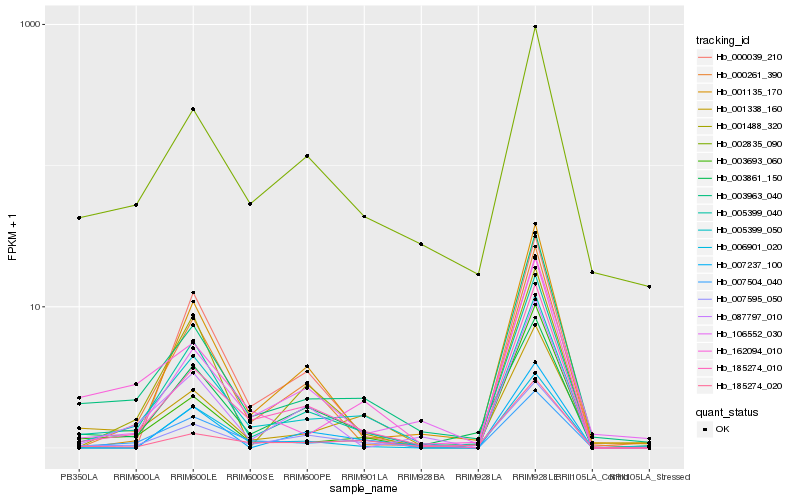
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005399_050 |
0.0 |
- |
- |
DNA-directed RNA polymerase subunit beta [Glycine soja] |
| 2 |
Hb_185274_010 |
0.1104388798 |
- |
- |
hypothetical protein ARALYDRAFT_330174 [Arabidopsis lyrata subsp. lyrata] |
| 3 |
Hb_007504_040 |
0.1159718381 |
- |
- |
hypothetical chloroplast RF2 [Fragaria iinumae] |
| 4 |
Hb_005399_040 |
0.1161673477 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 5 |
Hb_003963_040 |
0.1289835627 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 6 |
Hb_007595_050 |
0.1462252247 |
- |
- |
hypothetical chloroplast RF2 [Hevea brasiliensis] |
| 7 |
Hb_003861_150 |
0.1561061951 |
- |
- |
hypothetical protein JCGZ_15167 [Jatropha curcas] |
| 8 |
Hb_000261_390 |
0.1610893764 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 9 |
Hb_001338_160 |
0.1671886611 |
- |
- |
NADH dehydrogenase subunit 5 [Hevea brasiliensis] |
| 10 |
Hb_162094_010 |
0.1683227626 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |
| 11 |
Hb_001488_320 |
0.1747280146 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 12 |
Hb_106552_030 |
0.1753645054 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 13 |
Hb_185274_020 |
0.1759469951 |
- |
- |
Ycf2 (chloroplast) [Andrographis paniculata] |
| 14 |
Hb_006901_020 |
0.1761480708 |
- |
- |
PREDICTED: guard cell S-type anion channel SLAC1 [Jatropha curcas] |
| 15 |
Hb_087797_010 |
0.1762089925 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_001135_170 |
0.1771783035 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 17 |
Hb_002835_090 |
0.179323059 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 18 |
Hb_007237_100 |
0.1797880405 |
- |
- |
DNA-directed RNA polymerase beta chain, putative [Ricinus communis] |
| 19 |
Hb_000039_210 |
0.1805203005 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 20 |
Hb_003693_060 |
0.1808052557 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |