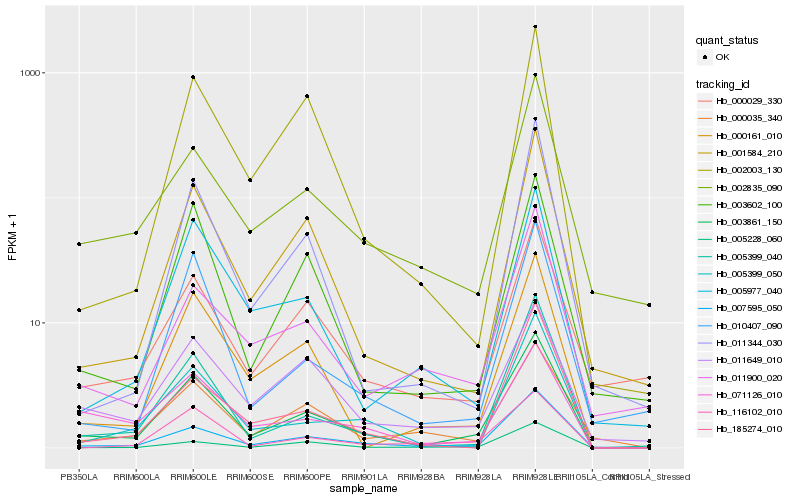
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003861_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_15167 [Jatropha curcas] |
| 2 |
Hb_007595_050 |
0.1024677602 |
- |
- |
hypothetical chloroplast RF2 [Hevea brasiliensis] |
| 3 |
Hb_005399_040 |
0.1422008993 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 4 |
Hb_001584_210 |
0.1469457045 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000161_010 |
0.1499127013 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_005228_060 |
0.1504508898 |
- |
- |
PREDICTED: uncharacterized protein LOC102604101 [Solanum tuberosum] |
| 7 |
Hb_002835_090 |
0.1520487719 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 8 |
Hb_005399_050 |
0.1561061951 |
- |
- |
DNA-directed RNA polymerase subunit beta [Glycine soja] |
| 9 |
Hb_000035_340 |
0.1584551225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003602_100 |
0.1599530725 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_116102_010 |
0.1599754626 |
- |
- |
hypothetical protein POPTR_0204s00240g [Populus trichocarpa] |
| 12 |
Hb_011649_010 |
0.1608474309 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 13 |
Hb_011900_020 |
0.1665631149 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000029_330 |
0.1672484987 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Jatropha curcas] |
| 15 |
Hb_071126_010 |
0.1679386849 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_005977_040 |
0.1697596824 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 17 |
Hb_185274_010 |
0.1713344066 |
- |
- |
hypothetical protein ARALYDRAFT_330174 [Arabidopsis lyrata subsp. lyrata] |
| 18 |
Hb_011344_030 |
0.1723232377 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_010407_090 |
0.1723972551 |
- |
- |
PREDICTED: aldo-keto reductase isoform X1 [Jatropha curcas] |
| 20 |
Hb_002003_130 |
0.1739985826 |
- |
- |
hypothetical protein CISIN_1g027843mg [Citrus sinensis] |