| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
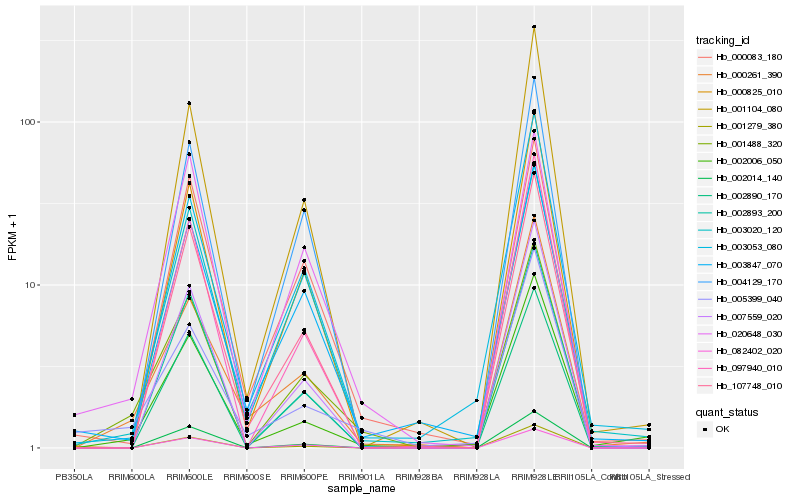
Hb_001488_320 |
0.0 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 2 |
Hb_002006_050 |
0.123753529 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 3 |
Hb_005399_040 |
0.124062316 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 4 |
Hb_000261_390 |
0.1301906425 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 5 |
Hb_003847_070 |
0.1315534486 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_020648_030 |
0.1337400387 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 7 |
Hb_000825_010 |
0.1351618316 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 8 |
Hb_107748_010 |
0.1363883625 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_002890_170 |
0.1364901146 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 10 |
Hb_003053_080 |
0.1366044807 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002893_200 |
0.1369565116 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_003020_120 |
0.1369704258 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 13 |
Hb_097940_010 |
0.1382801786 |
- |
- |
pollen protein Ole E I-like protein [Medicago truncatula] |
| 14 |
Hb_007559_020 |
0.1383129642 |
- |
- |
peptidylprolyl isomerase, putative [Ricinus communis] |
| 15 |
Hb_082402_020 |
0.138811732 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 16 |
Hb_001279_380 |
0.1392704112 |
- |
- |
PREDICTED: uncharacterized protein LOC105115824 isoform X1 [Populus euphratica] |
| 17 |
Hb_000083_180 |
0.139272079 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 18 |
Hb_002014_140 |
0.1393196649 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 19 |
Hb_004129_170 |
0.1395667794 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 20 |
Hb_001104_080 |
0.1396756535 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |