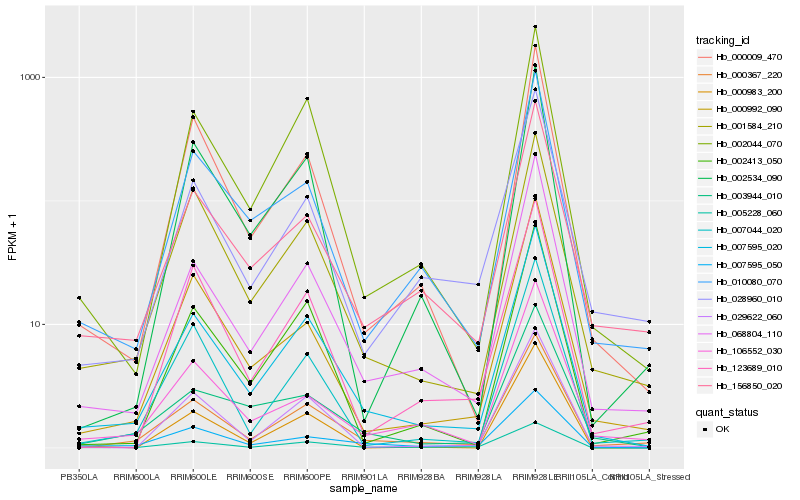
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007595_020 |
0.0 |
- |
- |
maturase K [Hevea brasiliensis] |
| 2 |
Hb_068804_110 |
0.0728294573 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000367_220 |
0.0886604108 |
- |
- |
hypothetical protein RCOM_1470580 [Ricinus communis] |
| 4 |
Hb_002044_070 |
0.0906567847 |
- |
- |
Chlorophyll a-b binding protein 3, chloroplastic [Theobroma cacao] |
| 5 |
Hb_005228_060 |
0.0961959609 |
- |
- |
PREDICTED: uncharacterized protein LOC102604101 [Solanum tuberosum] |
| 6 |
Hb_000009_470 |
0.1015422305 |
- |
- |
Oxygen-evolving enhancer protein 1, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_029622_060 |
0.1036250294 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_010080_070 |
0.1040772962 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 9 |
Hb_156850_020 |
0.1047888355 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |
| 10 |
Hb_002413_050 |
0.1071725436 |
- |
- |
hypothetical protein POPTR_0018s06150g [Populus trichocarpa] |
| 11 |
Hb_000992_090 |
0.1111066236 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 12 |
Hb_007595_050 |
0.1134409956 |
- |
- |
hypothetical chloroplast RF2 [Hevea brasiliensis] |
| 13 |
Hb_001584_210 |
0.1138349131 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 14 |
Hb_028960_010 |
0.1163913517 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 15 |
Hb_007044_020 |
0.1164107267 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_002534_090 |
0.1187338475 |
- |
- |
geranylgeranyl reductase [Hevea brasiliensis] |
| 17 |
Hb_003944_010 |
0.1202605987 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 18 |
Hb_000983_200 |
0.1224857214 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 19 |
Hb_106552_030 |
0.1227570883 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 20 |
Hb_123689_010 |
0.1234393895 |
- |
- |
PREDICTED: uncharacterized protein At1g18480 [Jatropha curcas] |