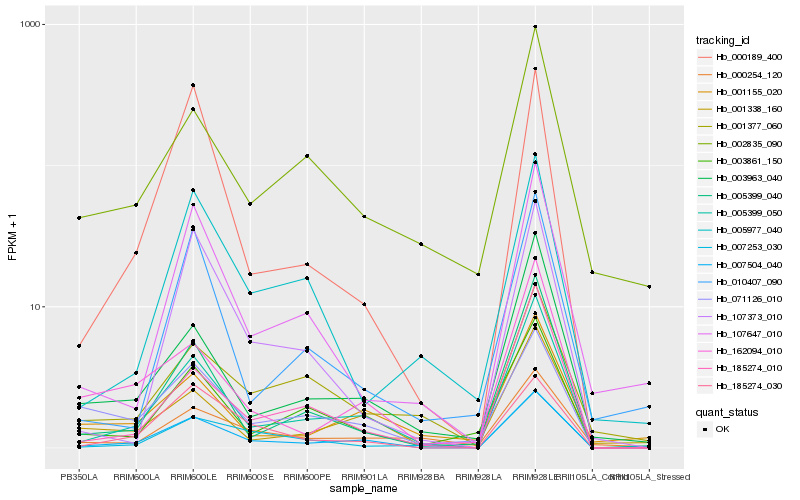
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007504_040 |
0.0 |
- |
- |
hypothetical chloroplast RF2 [Fragaria iinumae] |
| 2 |
Hb_005399_050 |
0.1159718381 |
- |
- |
DNA-directed RNA polymerase subunit beta [Glycine soja] |
| 3 |
Hb_185274_030 |
0.1715760356 |
- |
- |
hypothetical protein JCGZ_15170 [Jatropha curcas] |
| 4 |
Hb_001338_160 |
0.1744986865 |
- |
- |
NADH dehydrogenase subunit 5 [Hevea brasiliensis] |
| 5 |
Hb_005399_040 |
0.1776638648 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 6 |
Hb_107647_010 |
0.1845940202 |
- |
- |
PREDICTED: uncharacterized protein ycf39 [Jatropha curcas] |
| 7 |
Hb_001155_020 |
0.1846176829 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 2 [Theobroma cacao] |
| 8 |
Hb_003963_040 |
0.1868626214 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 9 |
Hb_185274_010 |
0.1872401944 |
- |
- |
hypothetical protein ARALYDRAFT_330174 [Arabidopsis lyrata subsp. lyrata] |
| 10 |
Hb_162094_010 |
0.1874906641 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |
| 11 |
Hb_003861_150 |
0.1881471945 |
- |
- |
hypothetical protein JCGZ_15167 [Jatropha curcas] |
| 12 |
Hb_000189_400 |
0.1904104838 |
- |
- |
hypothetical protein CathTA2_0066 [Caldalkalibacillus thermarum TA2.A1] |
| 13 |
Hb_000254_120 |
0.1912848232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007253_030 |
0.1917178102 |
- |
- |
PREDICTED: 33 kDa ribonucleoprotein, chloroplastic-like [Populus euphratica] |
| 15 |
Hb_071126_010 |
0.1958470811 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_002835_090 |
0.1965716293 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 17 |
Hb_001377_060 |
0.2011104201 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 18 |
Hb_010407_090 |
0.2016960426 |
- |
- |
PREDICTED: aldo-keto reductase isoform X1 [Jatropha curcas] |
| 19 |
Hb_107373_010 |
0.2048884442 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_005977_040 |
0.2064134443 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |