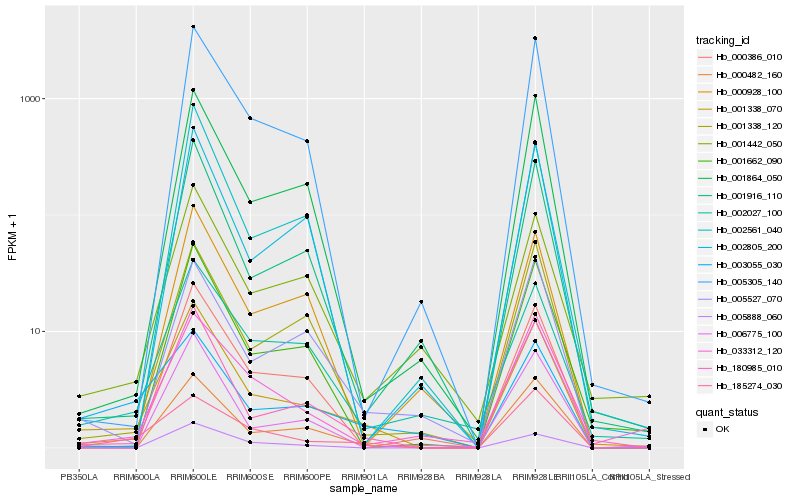
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001338_070 |
0.0 |
- |
- |
ORF126 [Jatropha curcas] |
| 2 |
Hb_180985_010 |
0.1322222265 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 1 [Theobroma cacao] |
| 3 |
Hb_000482_160 |
0.148232552 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 4 |
Hb_002027_100 |
0.154244724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001916_110 |
0.1564091797 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 6 |
Hb_185274_030 |
0.1595704146 |
- |
- |
hypothetical protein JCGZ_15170 [Jatropha curcas] |
| 7 |
Hb_005305_140 |
0.1600205663 |
- |
- |
unknown [Populus trichocarpa] |
| 8 |
Hb_001864_050 |
0.161763782 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 9 |
Hb_006775_100 |
0.1642517663 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000386_010 |
0.1652705629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001442_050 |
0.1655471822 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001662_090 |
0.1664237695 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_005527_070 |
0.1692131897 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |
| 14 |
Hb_001338_120 |
0.1694690497 |
- |
- |
hypothetical chloroplast RF68 (chloroplast) [Ipomoea batatas] |
| 15 |
Hb_005888_060 |
0.1739356651 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 16 |
Hb_003055_030 |
0.174612087 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 17 |
Hb_033312_120 |
0.1764903098 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000928_100 |
0.1781105535 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 19 |
Hb_002561_040 |
0.1781224628 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 20 |
Hb_002805_200 |
0.1790512976 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |