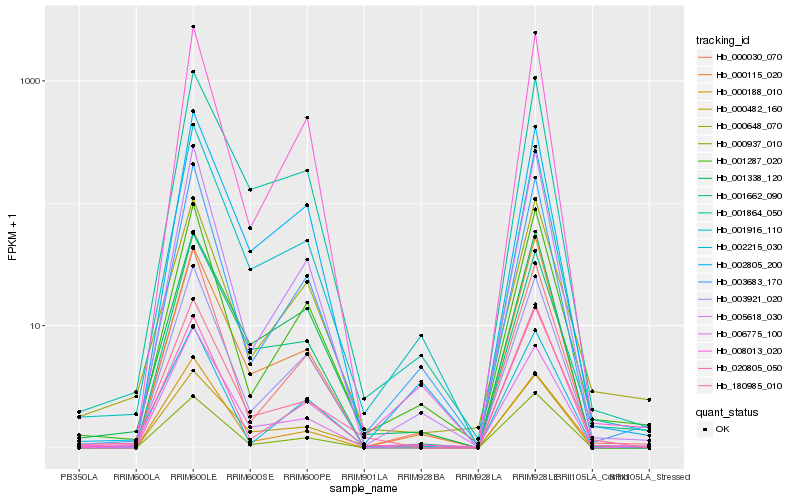
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_180985_010 |
0.0 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 1 [Theobroma cacao] |
| 2 |
Hb_000188_010 |
0.0946387795 |
- |
- |
PREDICTED: cytochrome P450 94C1-like [Jatropha curcas] |
| 3 |
Hb_000482_160 |
0.0998649647 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 4 |
Hb_001864_050 |
0.1074855564 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 5 |
Hb_001916_110 |
0.1080407554 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 6 |
Hb_003921_020 |
0.1105916798 |
- |
- |
hypothetical protein JCGZ_10628 [Jatropha curcas] |
| 7 |
Hb_001287_020 |
0.1107125144 |
- |
- |
SulA [Manihot esculenta] |
| 8 |
Hb_005618_030 |
0.1109289708 |
- |
- |
PREDICTED: cytochrome P450 734A1-like [Jatropha curcas] |
| 9 |
Hb_001662_090 |
0.1111222946 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_006775_100 |
0.1123677037 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_000937_010 |
0.113192658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000648_070 |
0.1151372702 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 13 |
Hb_000030_070 |
0.1160076902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002805_200 |
0.1197423911 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 15 |
Hb_003683_170 |
0.1213115368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001338_120 |
0.1226197678 |
- |
- |
hypothetical chloroplast RF68 (chloroplast) [Ipomoea batatas] |
| 17 |
Hb_020805_050 |
0.1229266123 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 18 |
Hb_008013_020 |
0.1239067777 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000115_020 |
0.1239083078 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 20 |
Hb_002215_030 |
0.1246852592 |
- |
- |
PREDICTED: uncharacterized protein LOC105640840 isoform X1 [Jatropha curcas] |