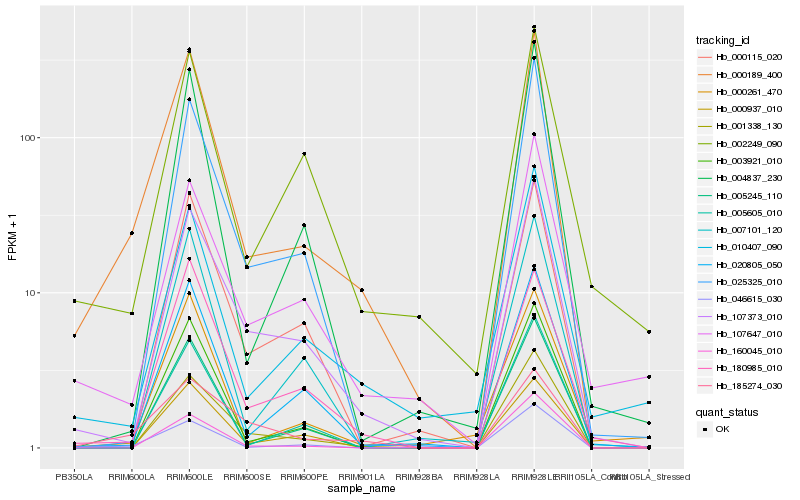
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_400 |
0.0 |
- |
- |
hypothetical protein CathTA2_0066 [Caldalkalibacillus thermarum TA2.A1] |
| 2 |
Hb_046615_030 |
0.116617755 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 3 |
Hb_001338_130 |
0.1433854348 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 4 |
Hb_107373_010 |
0.1436201237 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_020805_050 |
0.1451347738 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 6 |
Hb_003921_010 |
0.1509344702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_180985_010 |
0.1524537299 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 1 [Theobroma cacao] |
| 8 |
Hb_005245_110 |
0.1570656297 |
- |
- |
hypothetical protein CISIN_1g0225141mg [Citrus sinensis] |
| 9 |
Hb_010407_090 |
0.157231363 |
- |
- |
PREDICTED: aldo-keto reductase isoform X1 [Jatropha curcas] |
| 10 |
Hb_004837_230 |
0.1595331342 |
- |
- |
unknown [Populus trichocarpa] |
| 11 |
Hb_025325_010 |
0.1611912676 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 12 |
Hb_000115_020 |
0.1615806458 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 13 |
Hb_000261_470 |
0.164826933 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 14 |
Hb_160045_010 |
0.1671095946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005605_010 |
0.1671651054 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 16 |
Hb_002249_090 |
0.1674843562 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000937_010 |
0.1675216187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_185274_030 |
0.1677317099 |
- |
- |
hypothetical protein JCGZ_15170 [Jatropha curcas] |
| 19 |
Hb_007101_120 |
0.1681451213 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 20 |
Hb_107647_010 |
0.1692463921 |
- |
- |
PREDICTED: uncharacterized protein ycf39 [Jatropha curcas] |