| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
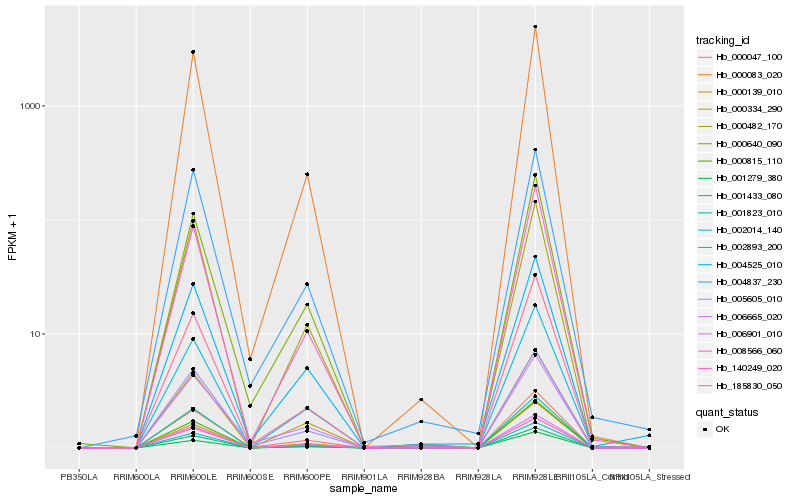
Hb_001433_080 |
0.0 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 2 |
Hb_140249_020 |
0.0316913728 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 3 |
Hb_000047_100 |
0.0435760338 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006901_010 |
0.0709127524 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 5 |
Hb_000083_020 |
0.0736634541 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 6 |
Hb_002014_140 |
0.0737256282 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 7 |
Hb_002893_200 |
0.0782470395 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000482_170 |
0.0806618443 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 9 |
Hb_004525_010 |
0.0837117758 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 10 |
Hb_005605_010 |
0.0850725051 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 11 |
Hb_000815_110 |
0.085787911 |
- |
- |
RecName: Full=Casparian strip membrane protein 4; Short=RcCASP4 [Ricinus communis] |
| 12 |
Hb_001279_380 |
0.0863131954 |
- |
- |
PREDICTED: uncharacterized protein LOC105115824 isoform X1 [Populus euphratica] |
| 13 |
Hb_008566_060 |
0.0866841658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004837_230 |
0.0869885671 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_000640_090 |
0.0900900282 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_000334_290 |
0.0918473523 |
- |
- |
monovalent cation:proton antiporter, putative [Ricinus communis] |
| 17 |
Hb_006665_020 |
0.0928008041 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 18 |
Hb_185830_050 |
0.0933320301 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 19 |
Hb_000139_010 |
0.094026298 |
- |
- |
- |
| 20 |
Hb_001823_010 |
0.0940522531 |
- |
- |
PREDICTED: ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |