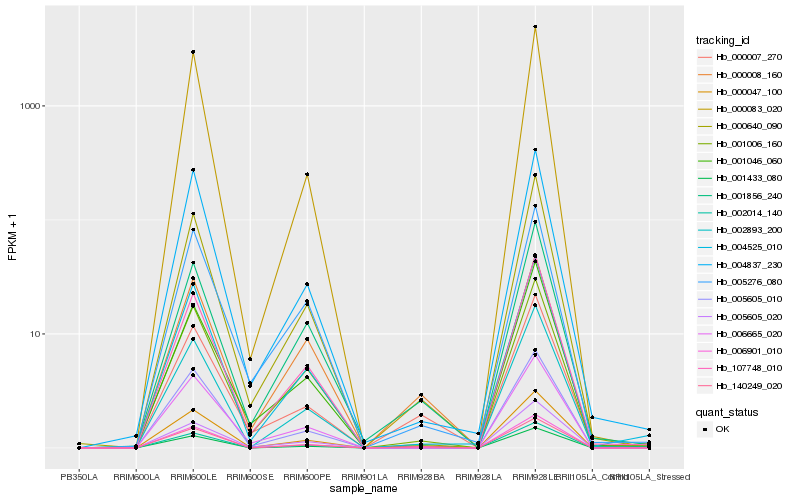
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000047_100 |
0.0 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001433_080 |
0.0435760338 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 3 |
Hb_140249_020 |
0.0502843978 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 4 |
Hb_006665_020 |
0.0712906377 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 5 |
Hb_000007_270 |
0.0760564457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001856_240 |
0.0769662918 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 7 |
Hb_001006_160 |
0.0777490436 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_000640_090 |
0.0796025795 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_004525_010 |
0.0804800755 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 10 |
Hb_107748_010 |
0.0835118879 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_002893_200 |
0.0837394438 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_005605_010 |
0.0841203181 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 13 |
Hb_002014_140 |
0.0851693296 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 14 |
Hb_006901_010 |
0.0857169509 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 15 |
Hb_004837_230 |
0.0865418626 |
- |
- |
unknown [Populus trichocarpa] |
| 16 |
Hb_000083_020 |
0.0872929836 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 17 |
Hb_005605_020 |
0.0915726967 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 18 |
Hb_005276_080 |
0.0926738892 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000008_160 |
0.0929425118 |
- |
- |
carboxy-lyase, putative [Ricinus communis] |
| 20 |
Hb_001046_060 |
0.0931792254 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |