| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
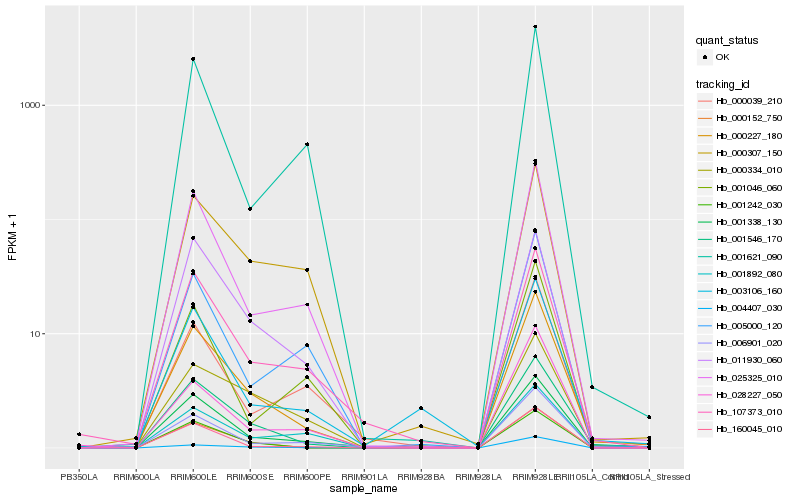
Hb_000227_180 |
0.0 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001546_170 |
0.0564156966 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_025325_010 |
0.0983387324 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 4 |
Hb_001338_130 |
0.1061498552 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 5 |
Hb_001242_030 |
0.1140220434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_160045_010 |
0.1156383092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006901_020 |
0.1195661041 |
- |
- |
PREDICTED: guard cell S-type anion channel SLAC1 [Jatropha curcas] |
| 8 |
Hb_107373_010 |
0.1222511783 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000307_150 |
0.1350973786 |
- |
- |
PREDICTED: thioredoxin-like protein CDSP32, chloroplastic [Jatropha curcas] |
| 10 |
Hb_011930_060 |
0.13564893 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 11 |
Hb_003106_160 |
0.1358407551 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 12 |
Hb_005000_120 |
0.1420233182 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 13 |
Hb_000334_010 |
0.1434703543 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 14 |
Hb_028227_050 |
0.1472732057 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_000039_210 |
0.1483413163 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 16 |
Hb_001892_080 |
0.1483453439 |
- |
- |
Clavata3/esr-related 16, putative [Theobroma cacao] |
| 17 |
Hb_001621_090 |
0.1486081609 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 18 |
Hb_000152_750 |
0.1517482721 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 19 |
Hb_001046_060 |
0.1553977799 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 20 |
Hb_004407_030 |
0.1564832108 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |