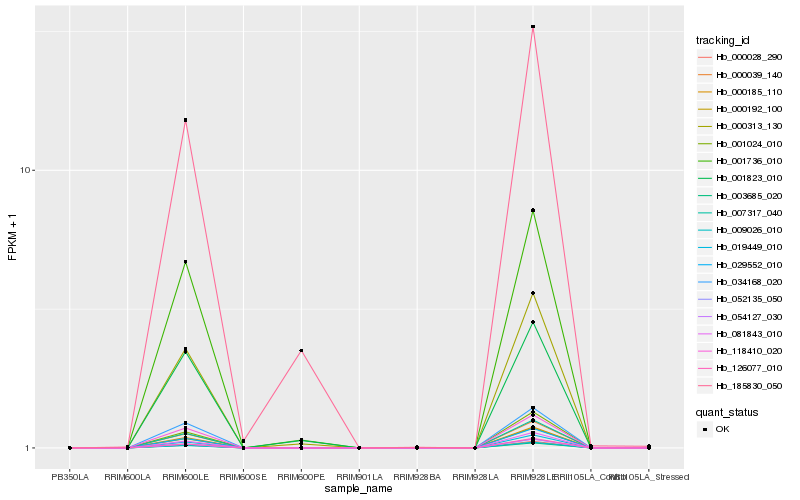
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000313_130 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_001736_010 |
0.0414478287 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_007317_040 |
0.0660613814 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 7 [Jatropha curcas] |
| 4 |
Hb_000185_110 |
0.0662972807 |
- |
- |
hypothetical protein JCGZ_11437 [Jatropha curcas] |
| 5 |
Hb_000039_140 |
0.0663220807 |
- |
- |
nuclease, putative [Ricinus communis] |
| 6 |
Hb_029552_010 |
0.0665883014 |
- |
- |
hypothetical protein JCGZ_08684 [Jatropha curcas] |
| 7 |
Hb_019449_010 |
0.0689018861 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_000192_100 |
0.0690268806 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 9 |
Hb_185830_050 |
0.0695262839 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 10 |
Hb_054127_030 |
0.0710232274 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 11 |
Hb_081843_010 |
0.071437865 |
- |
- |
PREDICTED: uncharacterized protein LOC104880938 [Vitis vinifera] |
| 12 |
Hb_118410_020 |
0.072785981 |
- |
- |
PREDICTED: UDP-glycosyltransferase 76C4 [Vitis vinifera] |
| 13 |
Hb_009026_010 |
0.0728300839 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 14 |
Hb_052135_050 |
0.0731163381 |
- |
- |
PREDICTED: probable pectinesterase 67 [Jatropha curcas] |
| 15 |
Hb_001823_010 |
0.0745555368 |
- |
- |
PREDICTED: ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 16 |
Hb_000028_290 |
0.0745720257 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 17 |
Hb_001024_010 |
0.0746083431 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 18 |
Hb_034168_020 |
0.0750219357 |
- |
- |
putative uncharacterized protein (mitochondrion) [Hevea brasiliensis] |
| 19 |
Hb_126077_010 |
0.0751514507 |
- |
- |
- |
| 20 |
Hb_003685_020 |
0.0764708856 |
- |
- |
PREDICTED: laccase-14-like [Fragaria vesca subsp. vesca] |