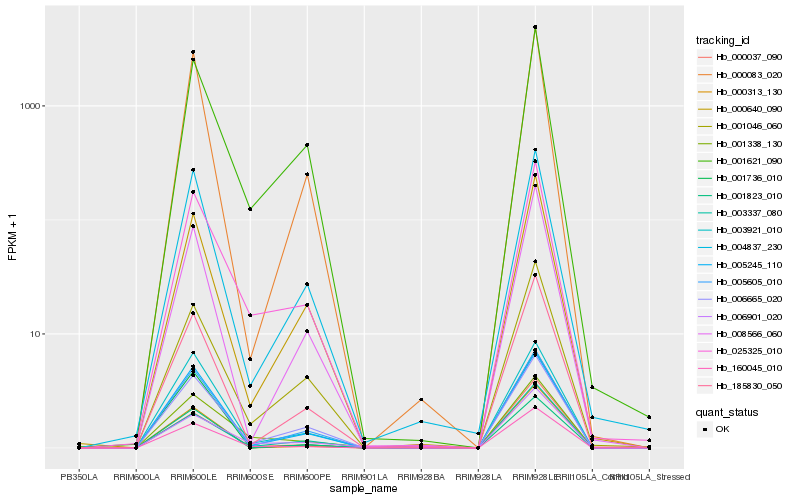
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_160045_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_025325_010 |
0.0697993448 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 3 |
Hb_185830_050 |
0.0867920584 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 4 |
Hb_000313_130 |
0.0874029574 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_001736_010 |
0.0945842807 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_000083_020 |
0.0965700213 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 7 |
Hb_005245_110 |
0.0968708228 |
- |
- |
hypothetical protein CISIN_1g0225141mg [Citrus sinensis] |
| 8 |
Hb_005605_010 |
0.0969047574 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 9 |
Hb_003337_080 |
0.0974784655 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 10 |
Hb_000037_090 |
0.0974999598 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 11 |
Hb_003921_010 |
0.0983499804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006901_020 |
0.0994034504 |
- |
- |
PREDICTED: guard cell S-type anion channel SLAC1 [Jatropha curcas] |
| 13 |
Hb_000640_090 |
0.1020572794 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_001823_010 |
0.1024495407 |
- |
- |
PREDICTED: ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 15 |
Hb_008566_060 |
0.1026065604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001621_090 |
0.1057965562 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 17 |
Hb_004837_230 |
0.106921001 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_001046_060 |
0.1086595393 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 19 |
Hb_001338_130 |
0.1106458355 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 20 |
Hb_006665_020 |
0.1114939013 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |