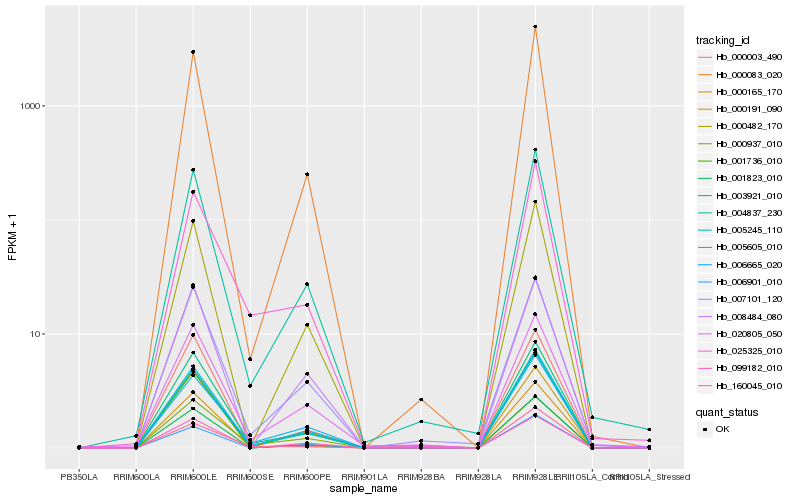
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003921_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005245_110 |
0.0272111649 |
- |
- |
hypothetical protein CISIN_1g0225141mg [Citrus sinensis] |
| 3 |
Hb_005605_010 |
0.0568755257 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 4 |
Hb_004837_230 |
0.0634960591 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_000003_490 |
0.0651008354 |
- |
- |
PREDICTED: uncharacterized protein LOC105631559 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001823_010 |
0.0681048249 |
- |
- |
PREDICTED: ribosome-inactivating protein cucurmosin-like [Jatropha curcas] |
| 7 |
Hb_000191_090 |
0.069251738 |
transcription factor |
TF Family: MYB |
GHMYB10, putative [Theobroma cacao] |
| 8 |
Hb_000083_020 |
0.0726181643 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 9 |
Hb_007101_120 |
0.0735596223 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 10 |
Hb_000482_170 |
0.0758704338 |
transcription factor |
TF Family: C2C2-GATA |
GATA nirate-inducible carbon-metabolism involved protein [Populus nigra x Populus x canadensis] |
| 11 |
Hb_000937_010 |
0.0863174371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_020805_050 |
0.0888680125 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 13 |
Hb_006665_020 |
0.089602521 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 14 |
Hb_099182_010 |
0.0915089692 |
- |
- |
PREDICTED: uncharacterized protein LOC101214783 [Cucumis sativus] |
| 15 |
Hb_000165_170 |
0.0944371584 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 16 |
Hb_160045_010 |
0.0983499804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006901_010 |
0.0986055715 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 18 |
Hb_008484_080 |
0.1006230532 |
- |
- |
PREDICTED: crocetin glucosyltransferase, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_001736_010 |
0.102655432 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_025325_010 |
0.1026731987 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |