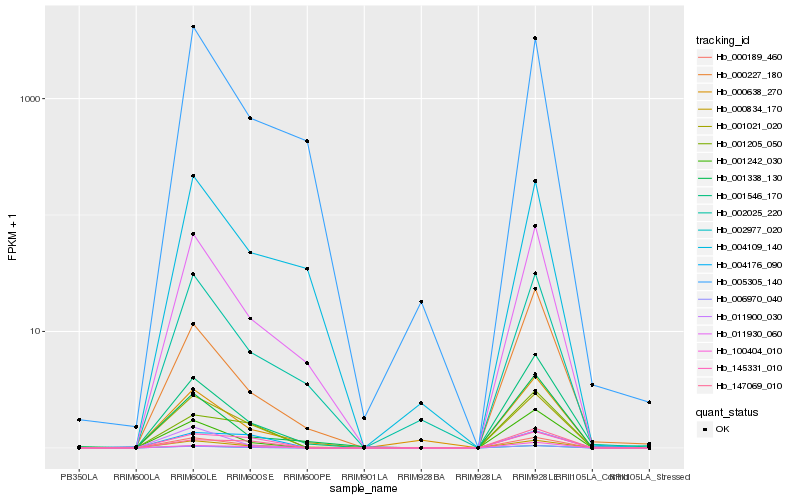
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_460 |
0.0 |
- |
- |
- |
| 2 |
Hb_001021_020 |
0.0269138847 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 3 |
Hb_002977_020 |
0.0343160774 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 1.8-like [Nelumbo nucifera] |
| 4 |
Hb_011900_030 |
0.1173473674 |
- |
- |
hypothetical protein JCGZ_13686 [Jatropha curcas] |
| 5 |
Hb_001242_030 |
0.1216877299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011930_060 |
0.1330311023 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 7 |
Hb_145331_010 |
0.1441238618 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0016s07690g [Populus trichocarpa] |
| 8 |
Hb_000834_170 |
0.1547260396 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 9 |
Hb_001546_170 |
0.1585179106 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_100404_010 |
0.1613234002 |
- |
- |
Cysteine-rich receptor-like protein kinase 29 [Glycine soja] |
| 11 |
Hb_002025_220 |
0.1621226674 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_001205_050 |
0.1633458108 |
- |
- |
- |
| 13 |
Hb_000638_270 |
0.1734411175 |
- |
- |
- |
| 14 |
Hb_005305_140 |
0.1787365166 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_004176_090 |
0.1787876949 |
- |
- |
Nuc-1 negative regulatory protein preg, putative [Ricinus communis] |
| 16 |
Hb_000227_180 |
0.1912392944 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_004109_140 |
0.1943290296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_147069_010 |
0.1945644649 |
- |
- |
Phosphate import ATP-binding PstB 2 [Gossypium arboreum] |
| 19 |
Hb_006970_040 |
0.1947925156 |
- |
- |
PREDICTED: uncharacterized protein LOC105650489 [Jatropha curcas] |
| 20 |
Hb_001338_130 |
0.1969002815 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |