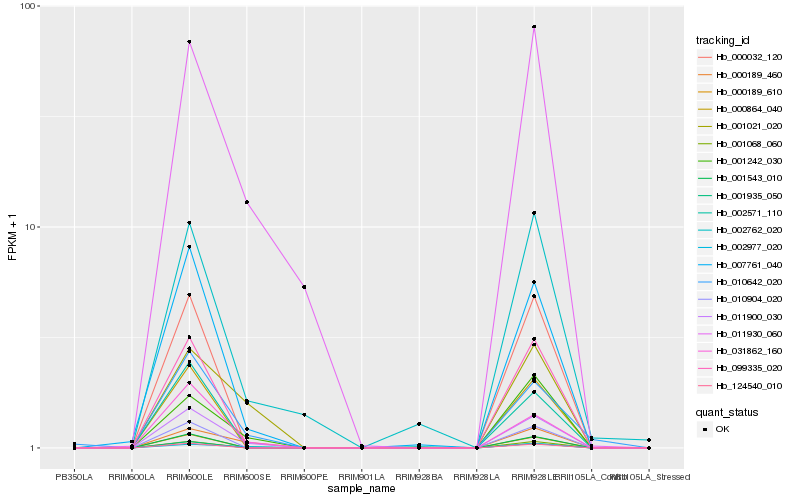
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011900_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_13686 [Jatropha curcas] |
| 2 |
Hb_007761_040 |
0.1081221348 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 3 |
Hb_000189_460 |
0.1173473674 |
- |
- |
- |
| 4 |
Hb_031862_160 |
0.117804615 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_002977_020 |
0.1299638271 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 1.8-like [Nelumbo nucifera] |
| 6 |
Hb_001021_020 |
0.1417152662 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 7 |
Hb_001242_030 |
0.1452806515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002571_110 |
0.1493634989 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 9 |
Hb_011930_060 |
0.14969703 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 10 |
Hb_010642_020 |
0.1612408562 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 2 [Theobroma cacao] |
| 11 |
Hb_000864_040 |
0.1622675758 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000189_610 |
0.1623638822 |
- |
- |
Late embryogenesis abundant protein D-34, putative [Ricinus communis] |
| 13 |
Hb_001543_010 |
0.1624103629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010904_020 |
0.1627803354 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 [Populus euphratica] |
| 15 |
Hb_124540_010 |
0.1629816582 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1-like [Populus euphratica] |
| 16 |
Hb_001935_050 |
0.1631855376 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 [Jatropha curcas] |
| 17 |
Hb_099335_020 |
0.1692779155 |
- |
- |
hypothetical protein ARALYDRAFT_905869 [Arabidopsis lyrata subsp. lyrata] |
| 18 |
Hb_002762_020 |
0.1697181661 |
- |
- |
hypothetical protein VITISV_043202 [Vitis vinifera] |
| 19 |
Hb_000032_120 |
0.1697527984 |
- |
- |
Bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase [Glycine soja] |
| 20 |
Hb_001068_060 |
0.1698309154 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |