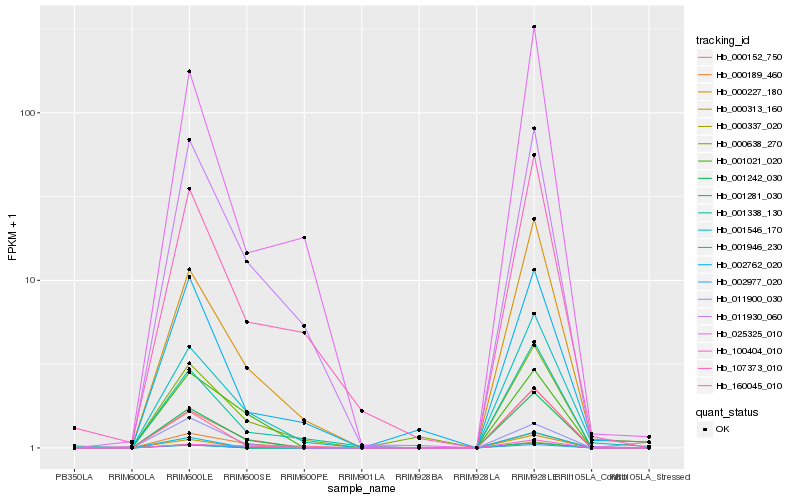
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001242_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002977_020 |
0.0932657118 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 1.8-like [Nelumbo nucifera] |
| 3 |
Hb_001546_170 |
0.1053989204 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000227_180 |
0.1140220434 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000189_460 |
0.1216877299 |
- |
- |
- |
| 6 |
Hb_001338_130 |
0.1322882249 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 7 |
Hb_011930_060 |
0.1336814219 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 8 |
Hb_160045_010 |
0.1362052011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001021_020 |
0.1400199136 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 10 |
Hb_000152_750 |
0.144648888 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 11 |
Hb_100404_010 |
0.1450932532 |
- |
- |
Cysteine-rich receptor-like protein kinase 29 [Glycine soja] |
| 12 |
Hb_011900_030 |
0.1452806515 |
- |
- |
hypothetical protein JCGZ_13686 [Jatropha curcas] |
| 13 |
Hb_025325_010 |
0.1487769234 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 14 |
Hb_000638_270 |
0.1583802067 |
- |
- |
- |
| 15 |
Hb_107373_010 |
0.1646633244 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_002762_020 |
0.1693442686 |
- |
- |
hypothetical protein VITISV_043202 [Vitis vinifera] |
| 17 |
Hb_000313_160 |
0.1709007838 |
- |
- |
BnaC02g20570D [Brassica napus] |
| 18 |
Hb_001946_230 |
0.1709296855 |
- |
- |
- |
| 19 |
Hb_000337_020 |
0.1709734966 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001281_030 |
0.1710389088 |
- |
- |
hypothetical protein POPTR_0010s18450g [Populus trichocarpa] |