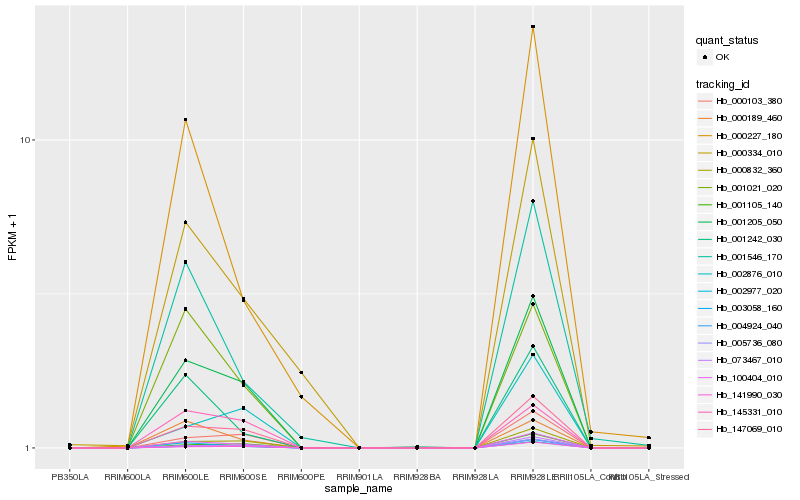
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_100404_010 |
0.0 |
- |
- |
Cysteine-rich receptor-like protein kinase 29 [Glycine soja] |
| 2 |
Hb_001205_050 |
0.0372986137 |
- |
- |
- |
| 3 |
Hb_147069_010 |
0.0551090817 |
- |
- |
Phosphate import ATP-binding PstB 2 [Gossypium arboreum] |
| 4 |
Hb_000832_360 |
0.0885041452 |
- |
- |
- |
| 5 |
Hb_073467_010 |
0.1154585636 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 6 |
Hb_000103_380 |
0.1172056186 |
- |
- |
PREDICTED: uncharacterized protein LOC104596624 [Nelumbo nucifera] |
| 7 |
Hb_003058_160 |
0.1208511329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004924_040 |
0.1210774838 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 9 |
Hb_001105_140 |
0.1249748283 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_005736_080 |
0.1277112404 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 11 |
Hb_141990_030 |
0.1292911462 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 12 |
Hb_002977_020 |
0.1334109344 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 1.8-like [Nelumbo nucifera] |
| 13 |
Hb_001242_030 |
0.1450932532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001546_170 |
0.1495179929 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_001021_020 |
0.1540769778 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 16 |
Hb_000227_180 |
0.1571260835 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000189_460 |
0.1613234002 |
- |
- |
- |
| 18 |
Hb_000334_010 |
0.163933761 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 19 |
Hb_002876_010 |
0.1681609494 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_145331_010 |
0.1703951308 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0016s07690g [Populus trichocarpa] |