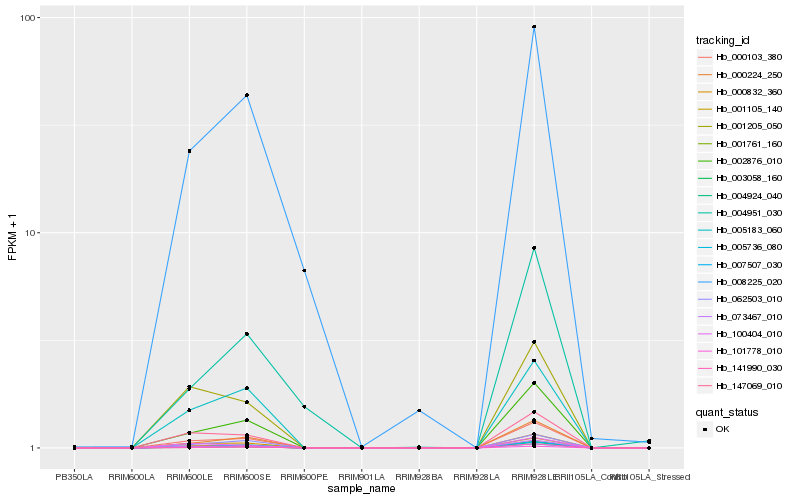
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_010 |
0.0 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000224_250 |
0.0246160898 |
- |
- |
hypothetical protein AMTR_s05400p00003230 [Amborella trichopoda] |
| 3 |
Hb_001105_140 |
0.0494637392 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_005736_080 |
0.0514025929 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 5 |
Hb_141990_030 |
0.0527922136 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 6 |
Hb_000103_380 |
0.0560031153 |
- |
- |
PREDICTED: uncharacterized protein LOC104596624 [Nelumbo nucifera] |
| 7 |
Hb_003058_160 |
0.0789424751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004924_040 |
0.0790933527 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 9 |
Hb_062503_010 |
0.0830384402 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_073467_010 |
0.0840739465 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 11 |
Hb_000832_360 |
0.0903755207 |
- |
- |
- |
| 12 |
Hb_005183_060 |
0.1177886546 |
- |
- |
hypothetical protein JCGZ_04271 [Jatropha curcas] |
| 13 |
Hb_101778_010 |
0.1179188549 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_147069_010 |
0.1257453063 |
- |
- |
Phosphate import ATP-binding PstB 2 [Gossypium arboreum] |
| 15 |
Hb_001761_160 |
0.1446475966 |
- |
- |
hypothetical protein VITISV_019364 [Vitis vinifera] |
| 16 |
Hb_007507_030 |
0.147085827 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 17 |
Hb_001205_050 |
0.1570125571 |
- |
- |
- |
| 18 |
Hb_008225_020 |
0.1587292708 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 19 |
Hb_100404_010 |
0.1681609494 |
- |
- |
Cysteine-rich receptor-like protein kinase 29 [Glycine soja] |
| 20 |
Hb_004951_030 |
0.1752574876 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |