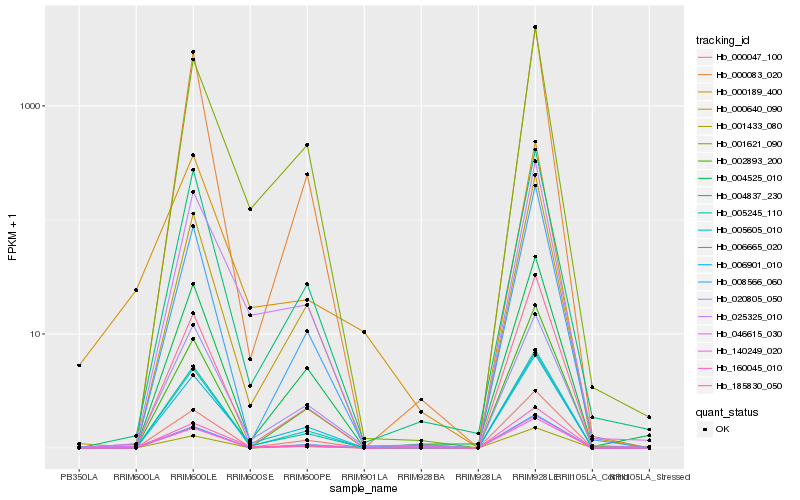
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_046615_030 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 2 |
Hb_000189_400 |
0.116617755 |
- |
- |
hypothetical protein CathTA2_0066 [Caldalkalibacillus thermarum TA2.A1] |
| 3 |
Hb_000047_100 |
0.1239431208 |
transcription factor |
TF Family: GRF |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004837_230 |
0.1280066156 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_006665_020 |
0.1303672453 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 6 |
Hb_001433_080 |
0.1313121103 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 7 |
Hb_005605_010 |
0.1321480909 |
- |
- |
hypothetical protein POPTR_0019s13220g [Populus trichocarpa] |
| 8 |
Hb_000640_090 |
0.1331513852 |
- |
- |
PREDICTED: calvin cycle protein CP12-2, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000083_020 |
0.1337805306 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 10 |
Hb_020805_050 |
0.1346194867 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 11 |
Hb_005245_110 |
0.1370128504 |
- |
- |
hypothetical protein CISIN_1g0225141mg [Citrus sinensis] |
| 12 |
Hb_185830_050 |
0.1372636311 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 [Jatropha curcas] |
| 13 |
Hb_025325_010 |
0.1402202492 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 14 |
Hb_140249_020 |
0.1402780956 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein CMB1-like [Jatropha curcas] |
| 15 |
Hb_004525_010 |
0.1407920614 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 16 |
Hb_160045_010 |
0.1412746617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001621_090 |
0.141930045 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 18 |
Hb_006901_010 |
0.1424326138 |
- |
- |
hypothetical protein JCGZ_20722 [Jatropha curcas] |
| 19 |
Hb_008566_060 |
0.1427716914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002893_200 |
0.1430784761 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |