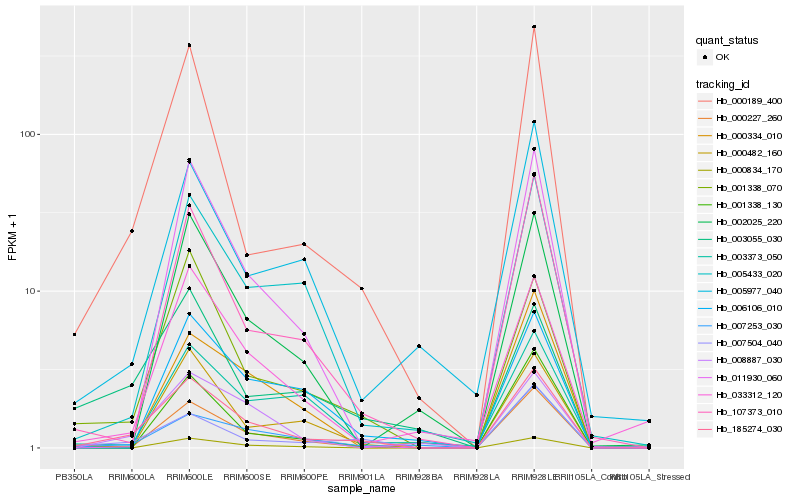
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185274_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_15170 [Jatropha curcas] |
| 2 |
Hb_001338_070 |
0.1595704146 |
- |
- |
ORF126 [Jatropha curcas] |
| 3 |
Hb_000189_400 |
0.1677317099 |
- |
- |
hypothetical protein CathTA2_0066 [Caldalkalibacillus thermarum TA2.A1] |
| 4 |
Hb_007504_040 |
0.1715760356 |
- |
- |
hypothetical chloroplast RF2 [Fragaria iinumae] |
| 5 |
Hb_007253_030 |
0.1725487718 |
- |
- |
PREDICTED: 33 kDa ribonucleoprotein, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_008887_030 |
0.176356952 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |
| 7 |
Hb_005433_020 |
0.1848766471 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 8 |
Hb_000227_260 |
0.1859888992 |
- |
- |
PREDICTED: RAN GTPase-activating protein 2 [Jatropha curcas] |
| 9 |
Hb_107373_010 |
0.186643997 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_003055_030 |
0.187629791 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |
| 11 |
Hb_011930_060 |
0.1915272734 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 12 |
Hb_033312_120 |
0.1921894022 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000834_170 |
0.1950093798 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 14 |
Hb_001338_130 |
0.1994955817 |
- |
- |
hypothetical protein MTR_4g129340 [Medicago truncatula] |
| 15 |
Hb_005977_040 |
0.2024498031 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 16 |
Hb_006106_010 |
0.2030113889 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_003373_050 |
0.2057232597 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 18 |
Hb_000482_160 |
0.2063560181 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 19 |
Hb_002025_220 |
0.2087467778 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000334_010 |
0.2097253684 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |