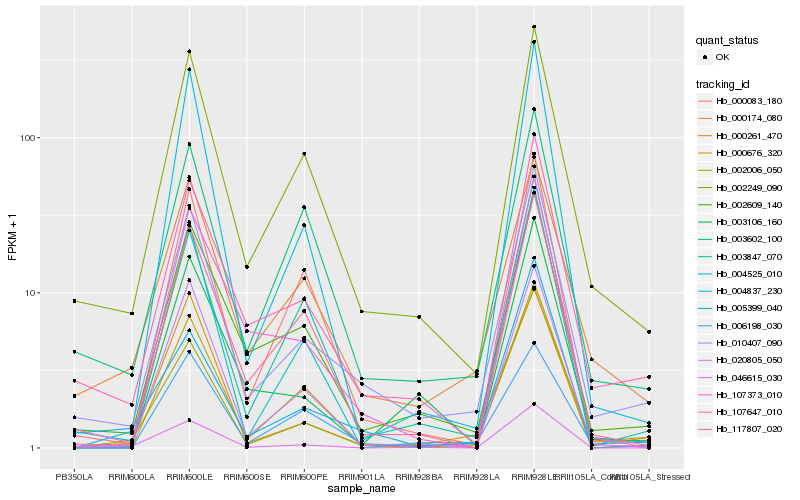
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010407_090 |
0.0 |
- |
- |
PREDICTED: aldo-keto reductase isoform X1 [Jatropha curcas] |
| 2 |
Hb_107647_010 |
0.0942405935 |
- |
- |
PREDICTED: uncharacterized protein ycf39 [Jatropha curcas] |
| 3 |
Hb_002249_090 |
0.1045864373 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002609_140 |
0.1167537066 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_002006_050 |
0.1300789157 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 6 |
Hb_000174_080 |
0.1304924169 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |
| 7 |
Hb_000261_470 |
0.1379753931 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 8 |
Hb_000676_320 |
0.1383918889 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 9 |
Hb_003847_070 |
0.1392954784 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_020805_050 |
0.1406355806 |
- |
- |
PREDICTED: MLO-like protein 9 [Jatropha curcas] |
| 11 |
Hb_004525_010 |
0.1411299696 |
- |
- |
late embryogenesis abundant, putative [Ricinus communis] |
| 12 |
Hb_003602_100 |
0.1420808978 |
- |
- |
Glutamyl-tRNA reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_004837_230 |
0.1435226217 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_000083_180 |
0.1440607825 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 15 |
Hb_117807_020 |
0.1444977432 |
- |
- |
methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_107373_010 |
0.1447219057 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_003106_160 |
0.1471592669 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 18 |
Hb_046615_030 |
0.1489949515 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 19 |
Hb_005399_040 |
0.1493565679 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 20 |
Hb_006198_030 |
0.1497079792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |