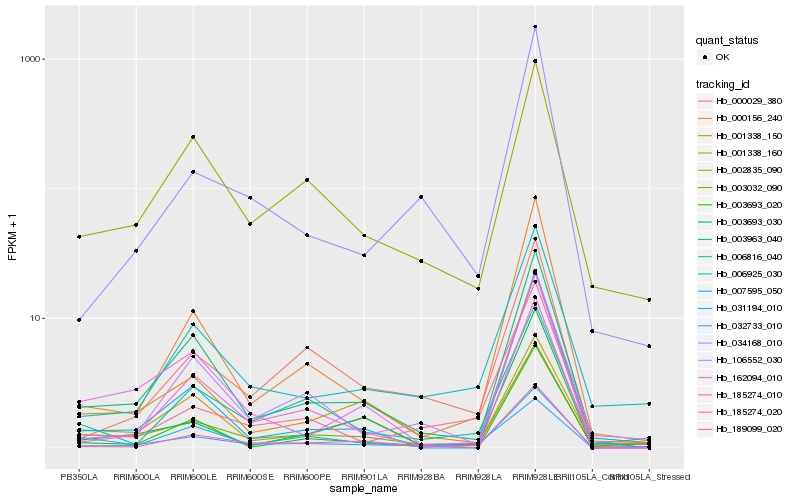
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003032_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 2 |
Hb_001338_150 |
0.0811339037 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 3 |
Hb_003963_040 |
0.0961915124 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 4 |
Hb_185274_020 |
0.1352939382 |
- |
- |
Ycf2 (chloroplast) [Andrographis paniculata] |
| 5 |
Hb_000156_240 |
0.1377575188 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 6 |
Hb_001338_160 |
0.1490060108 |
- |
- |
NADH dehydrogenase subunit 5 [Hevea brasiliensis] |
| 7 |
Hb_002835_090 |
0.1495197264 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 8 |
Hb_006816_040 |
0.1507571416 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 9 |
Hb_162094_010 |
0.1518743988 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |
| 10 |
Hb_007595_050 |
0.1585372068 |
- |
- |
hypothetical chloroplast RF2 [Hevea brasiliensis] |
| 11 |
Hb_006925_030 |
0.1588615865 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 12 |
Hb_003693_030 |
0.1591695221 |
- |
- |
NADH dehydrogenase subunit 7 [Hevea brasiliensis] |
| 13 |
Hb_032733_010 |
0.1600642599 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 14 |
Hb_003693_020 |
0.1631780174 |
- |
- |
Ycf1, partial (chloroplast) [Clusia rosea] |
| 15 |
Hb_000029_380 |
0.1650942181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_034168_010 |
0.1663948232 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 17 |
Hb_031194_010 |
0.1675006369 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Arachis hypogaea] |
| 18 |
Hb_185274_010 |
0.1682504095 |
- |
- |
hypothetical protein ARALYDRAFT_330174 [Arabidopsis lyrata subsp. lyrata] |
| 19 |
Hb_106552_030 |
0.1686916602 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 20 |
Hb_189099_020 |
0.1693097535 |
- |
- |
NADH dehydrogenase subunit 9 (mitochondrion) [Hevea brasiliensis] |