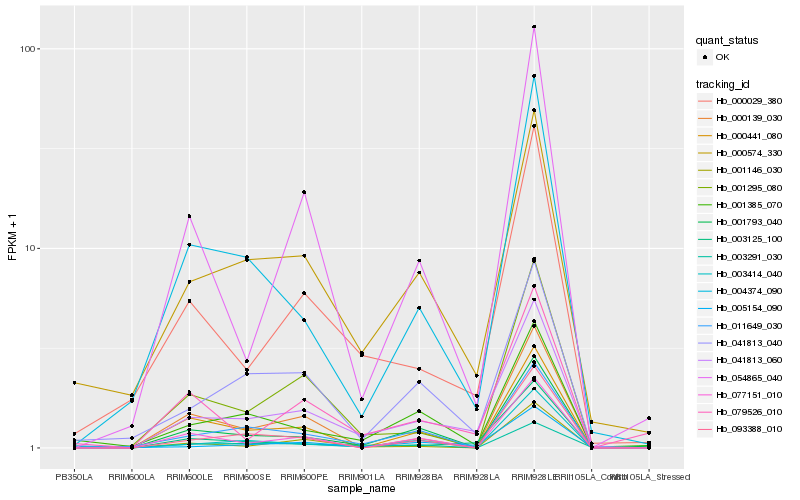
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041813_060 |
0.0 |
- |
- |
ribosomal protein L10 (mitochondrion) [Ricinus communis] |
| 2 |
Hb_000029_380 |
0.148200765 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000574_330 |
0.1578717422 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 4 |
Hb_003125_100 |
0.1596469454 |
- |
- |
NAD(P)-binding Rossmann-fold superfamily protein [Theobroma cacao] |
| 5 |
Hb_005154_090 |
0.1611248558 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_041813_040 |
0.1628869818 |
- |
- |
NADH2 dehydrogenase (ubiquinone) (EC 1.6.5.3) chain 5, truncated - sugar beet mitochondrion (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_001295_080 |
0.1692441467 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 8 |
Hb_004374_090 |
0.1699235078 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 9 |
Hb_003291_030 |
0.1766274721 |
- |
- |
Gag-pol protein, putative [Solanum demissum] |
| 10 |
Hb_001793_040 |
0.1808951719 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 11 |
Hb_001146_030 |
0.1811457616 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_093388_010 |
0.1814261791 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 13 |
Hb_003414_040 |
0.1814997165 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 14 |
Hb_054865_040 |
0.1819837183 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 15 |
Hb_011649_030 |
0.1825927711 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 16 |
Hb_079526_010 |
0.1829612148 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Populus euphratica] |
| 17 |
Hb_000441_080 |
0.184927649 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 18 |
Hb_077151_010 |
0.1858602011 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 19 |
Hb_001385_070 |
0.1860534632 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 20 |
Hb_000139_030 |
0.1871171728 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |