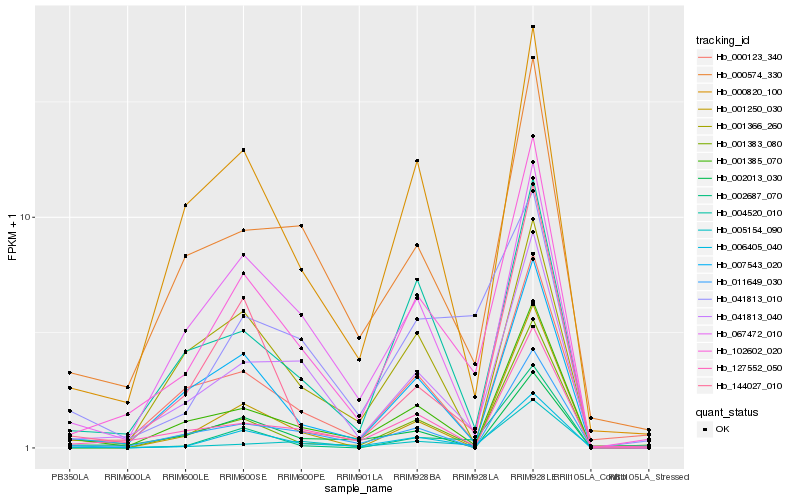
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_102602_020 |
0.0 |
- |
- |
hypothetical protein B456_001G166300 [Gossypium raimondii] |
| 2 |
Hb_041813_040 |
0.1268732106 |
- |
- |
NADH2 dehydrogenase (ubiquinone) (EC 1.6.5.3) chain 5, truncated - sugar beet mitochondrion (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_006405_040 |
0.1356118001 |
- |
- |
hypothetical protein VITISV_026680 [Vitis vinifera] |
| 4 |
Hb_001385_070 |
0.1450371221 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 5 |
Hb_127552_050 |
0.1460791276 |
- |
- |
hypothetical protein VITISV_030752 [Vitis vinifera] |
| 6 |
Hb_000820_100 |
0.1516600964 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 7 |
Hb_007543_020 |
0.1523336272 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 8 |
Hb_004520_010 |
0.1524008691 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 9 |
Hb_000123_340 |
0.1557994984 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 10 |
Hb_000574_330 |
0.1568637108 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 11 |
Hb_011649_030 |
0.165687166 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 12 |
Hb_001383_080 |
0.1686781078 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |
| 13 |
Hb_067472_010 |
0.1742434473 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 14 |
Hb_144027_010 |
0.1801742123 |
- |
- |
- |
| 15 |
Hb_001366_260 |
0.1825118973 |
- |
- |
T4.15 [Malus x robusta] |
| 16 |
Hb_041813_010 |
0.183725709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005154_090 |
0.1839936745 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_002687_070 |
0.1845255522 |
- |
- |
polyprotein [Ananas comosus] |
| 19 |
Hb_002013_030 |
0.1864287588 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 20 |
Hb_001250_030 |
0.1891811239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |