| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
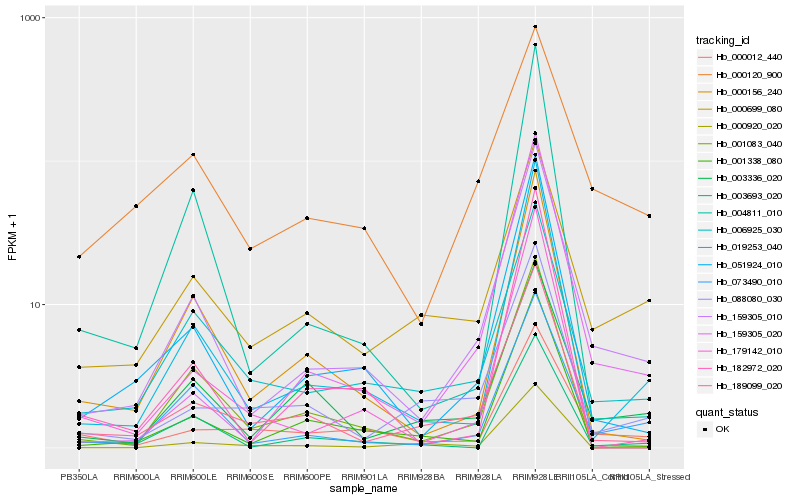
Hb_159305_010 |
0.0 |
- |
- |
rpl2 [Jatropha curcas] |
| 2 |
Hb_073490_010 |
0.1123513122 |
- |
- |
NAD(P)-linked oxidoreductase superfamily protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_051924_010 |
0.1278204805 |
- |
- |
BnaA01g34100D [Brassica napus] |
| 4 |
Hb_019253_040 |
0.1385077954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006925_030 |
0.1510233395 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 6 |
Hb_189099_020 |
0.1536186941 |
- |
- |
NADH dehydrogenase subunit 9 (mitochondrion) [Hevea brasiliensis] |
| 7 |
Hb_003336_020 |
0.1604416406 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS3, chloroplastic [Vitis vinifera] |
| 8 |
Hb_159305_020 |
0.1610152337 |
- |
- |
50S ribosomal protein L2-B [Medicago truncatula] |
| 9 |
Hb_001338_080 |
0.1618870346 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Tamarindus indica] |
| 10 |
Hb_000156_240 |
0.1628796742 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 11 |
Hb_003693_020 |
0.1691799236 |
- |
- |
Ycf1, partial (chloroplast) [Clusia rosea] |
| 12 |
Hb_000120_900 |
0.1731154618 |
- |
- |
catalase [Hevea brasiliensis] |
| 13 |
Hb_001083_040 |
0.1751700107 |
- |
- |
hypothetical protein EUGRSUZ_E00717 [Eucalyptus grandis] |
| 14 |
Hb_000699_080 |
0.1754415903 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_088080_030 |
0.1778462149 |
- |
- |
ribosomal protein S4 [Hevea brasiliensis] |
| 16 |
Hb_004811_010 |
0.1802487658 |
- |
- |
DNA-directed RNA polymerase subunit beta, partial [Medicago truncatula] |
| 17 |
Hb_182972_020 |
0.1807050612 |
- |
- |
cytochrome b6 [Gossypium barbadense] |
| 18 |
Hb_179142_010 |
0.1826783444 |
- |
- |
NADH dehydrogenase ND 2 subunit [Nicotiana tomentosiformis] |
| 19 |
Hb_000012_440 |
0.1829313089 |
- |
- |
PREDICTED: uncharacterized protein LOC104248335 [Nicotiana sylvestris] |
| 20 |
Hb_000920_020 |
0.1840167741 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |