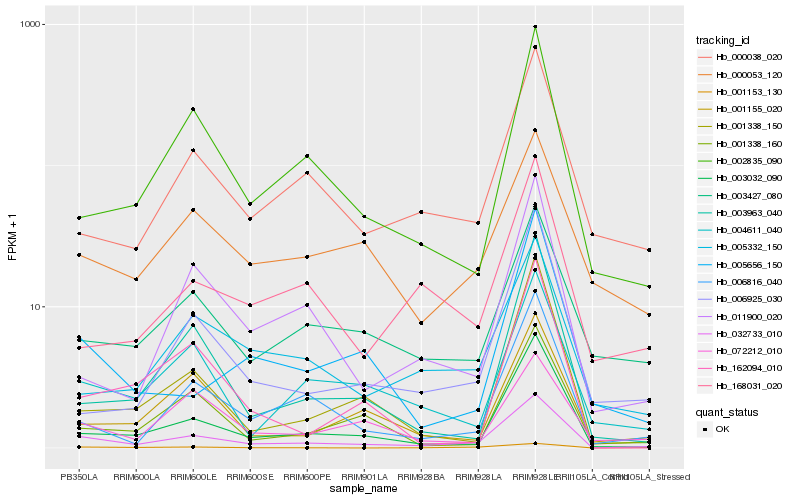
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032733_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 2 |
Hb_003032_090 |
0.1600642599 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 3 |
Hb_006816_040 |
0.178635966 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 4 |
Hb_002835_090 |
0.1881905128 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 5 |
Hb_004611_040 |
0.1908773087 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigD, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000038_020 |
0.1916708593 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001155_020 |
0.1990282119 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 2 [Theobroma cacao] |
| 8 |
Hb_003427_080 |
0.1998500252 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_011900_020 |
0.2011529624 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001153_130 |
0.201163045 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_005332_150 |
0.2013825233 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |
| 12 |
Hb_006925_030 |
0.202524015 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 13 |
Hb_001338_160 |
0.2039591984 |
- |
- |
NADH dehydrogenase subunit 5 [Hevea brasiliensis] |
| 14 |
Hb_003963_040 |
0.2047790049 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 15 |
Hb_001338_150 |
0.2052438132 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 16 |
Hb_000053_120 |
0.209974374 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_072212_010 |
0.2129448852 |
- |
- |
hypothetical chloroplast RF1 [Ficus sp. M. J. Moore 315] |
| 18 |
Hb_005656_150 |
0.2135498273 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 19 |
Hb_168031_020 |
0.2161860334 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 20 |
Hb_162094_010 |
0.2184709453 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |