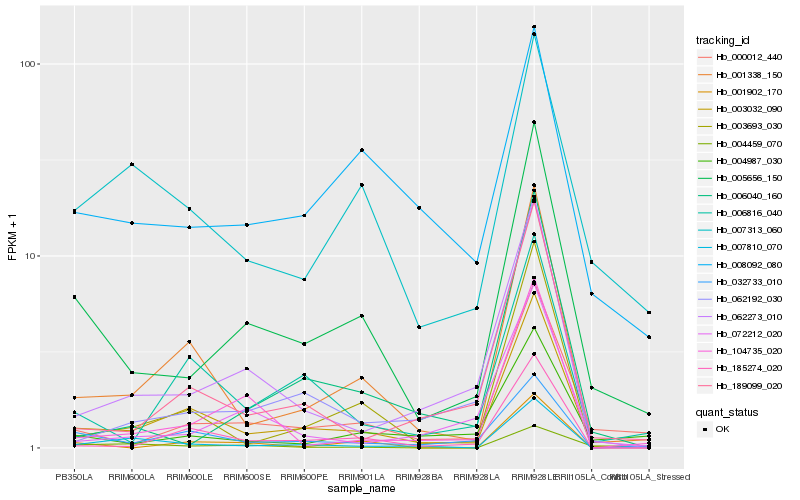
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005656_150 |
0.0 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 2 |
Hb_000012_440 |
0.1448398606 |
- |
- |
PREDICTED: uncharacterized protein LOC104248335 [Nicotiana sylvestris] |
| 3 |
Hb_003032_090 |
0.1799171308 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 4 |
Hb_004987_030 |
0.1971785849 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 5 |
Hb_001338_150 |
0.212133544 |
- |
- |
hypothetical chloroplast RF1 [Dillenia indica] |
| 6 |
Hb_032733_010 |
0.2135498273 |
- |
- |
hypothetical protein JCGZ_04542 [Jatropha curcas] |
| 7 |
Hb_008092_080 |
0.2162114586 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 8 |
Hb_001902_170 |
0.2165656344 |
- |
- |
PREDICTED: putative glucan endo-1,3-beta-glucosidase GVI [Nicotiana sylvestris] |
| 9 |
Hb_007313_060 |
0.2231577036 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |
| 10 |
Hb_006040_160 |
0.2237160634 |
- |
- |
ribosomal protein S12 (mitochondrion) [Hevea brasiliensis] |
| 11 |
Hb_062273_010 |
0.2326313356 |
- |
- |
ATP synthase F0 subunit 9 [Chara vulgaris] |
| 12 |
Hb_072212_020 |
0.2332479604 |
- |
- |
hypothetical protein B456_001G162300 [Gossypium raimondii] |
| 13 |
Hb_003693_030 |
0.234800457 |
- |
- |
NADH dehydrogenase subunit 7 [Hevea brasiliensis] |
| 14 |
Hb_104735_020 |
0.2372660104 |
- |
- |
ATPase subunit 4 (mitochondrion) [Hevea brasiliensis] |
| 15 |
Hb_062192_030 |
0.2375197891 |
- |
- |
ribosomal protein S1 (mitochondrion) [Hevea brasiliensis] |
| 16 |
Hb_006816_040 |
0.2381028359 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g48910-like [Jatropha curcas] |
| 17 |
Hb_185274_020 |
0.2404776034 |
- |
- |
Ycf2 (chloroplast) [Andrographis paniculata] |
| 18 |
Hb_189099_020 |
0.2407388512 |
- |
- |
NADH dehydrogenase subunit 9 (mitochondrion) [Hevea brasiliensis] |
| 19 |
Hb_004459_070 |
0.2461885647 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 20 |
Hb_007810_070 |
0.2472009552 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |