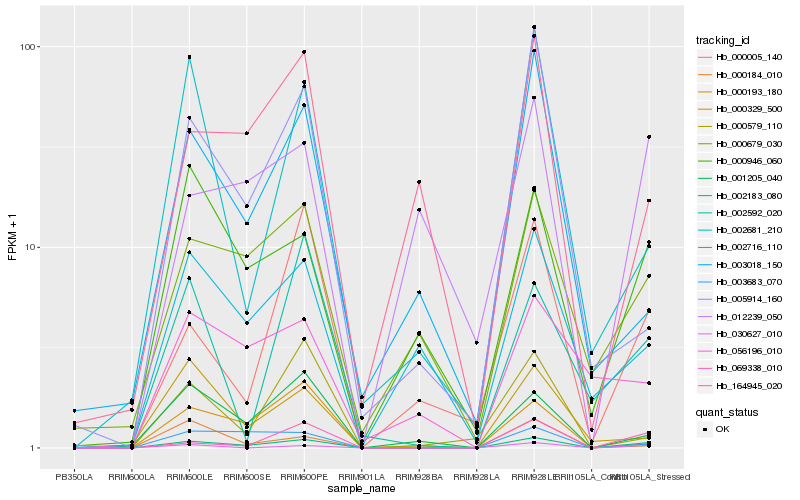
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002183_080 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_069338_010 |
0.2173244821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_030627_010 |
0.2194163365 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 4 |
Hb_000679_030 |
0.2257472256 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5 [Jatropha curcas] |
| 5 |
Hb_000946_060 |
0.2268019988 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_003683_070 |
0.2291189942 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_001205_040 |
0.236436437 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000184_010 |
0.2447725283 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 9 |
Hb_002716_110 |
0.2486111915 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 10 |
Hb_000005_140 |
0.2534806952 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 11 |
Hb_164945_020 |
0.2628457762 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 12 |
Hb_012239_050 |
0.2633147718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000329_500 |
0.2714492166 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002592_020 |
0.2718234286 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002681_210 |
0.2724938241 |
- |
- |
PREDICTED: uncharacterized protein LOC105633686 [Jatropha curcas] |
| 16 |
Hb_000193_180 |
0.2765313361 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB28-like [Jatropha curcas] |
| 17 |
Hb_000579_110 |
0.2768144808 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 18 |
Hb_056196_010 |
0.278759094 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 19 |
Hb_003018_150 |
0.2788935366 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 20 |
Hb_005914_160 |
0.2808898411 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |