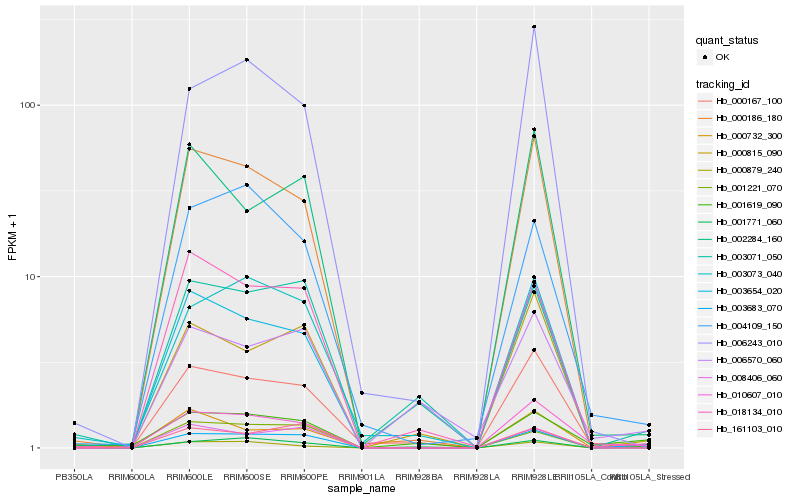
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003683_070 |
0.0 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 2 |
Hb_010607_010 |
0.1638288538 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 3 |
Hb_000879_240 |
0.1645130647 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631008 [Jatropha curcas] |
| 4 |
Hb_003654_020 |
0.1691674217 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_001771_060 |
0.1692199002 |
- |
- |
hypothetical protein B456_012G144100 [Gossypium raimondii] |
| 6 |
Hb_000815_090 |
0.17716328 |
- |
- |
PREDICTED: uncharacterized protein LOC105630464 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000186_180 |
0.1807329071 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 8 |
Hb_000732_300 |
0.1841602317 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003071_050 |
0.1876018695 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 2 [Hevea brasiliensis] |
| 10 |
Hb_008406_060 |
0.1889581945 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 11 |
Hb_161103_010 |
0.1912581728 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_018134_010 |
0.1926282224 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001619_090 |
0.1926423057 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_003073_040 |
0.1990561877 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 15 |
Hb_000167_100 |
0.2020126393 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_001221_070 |
0.2030527673 |
- |
- |
PREDICTED: calcium-binding protein CML38-like [Jatropha curcas] |
| 17 |
Hb_004109_150 |
0.2033710391 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 18 |
Hb_006243_010 |
0.2038275984 |
- |
- |
PREDICTED: cell wall protein IFF6 [Populus euphratica] |
| 19 |
Hb_002284_160 |
0.2045355232 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_006570_060 |
0.2065425381 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |