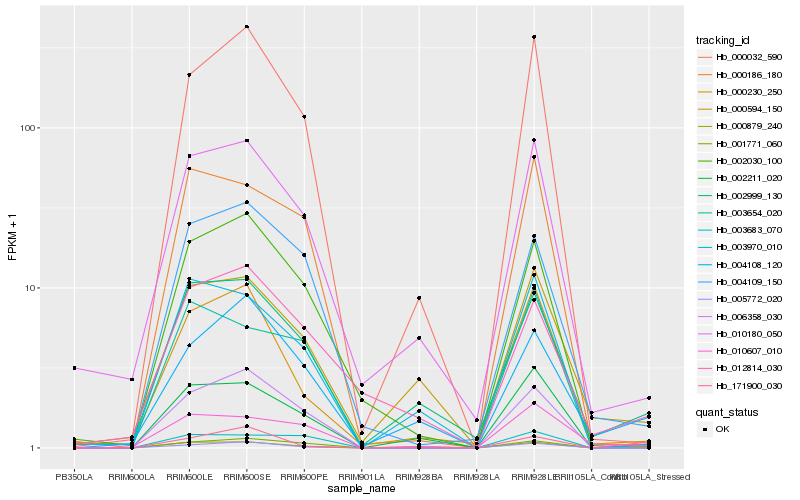
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000879_240 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631008 [Jatropha curcas] |
| 2 |
Hb_003683_070 |
0.1645130647 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 3 |
Hb_002999_130 |
0.1784786185 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 4 |
Hb_001771_060 |
0.1843210154 |
- |
- |
hypothetical protein B456_012G144100 [Gossypium raimondii] |
| 5 |
Hb_000186_180 |
0.2096631032 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 6 |
Hb_010607_010 |
0.2106335462 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 7 |
Hb_000230_250 |
0.2112098245 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004108_120 |
0.2115441594 |
- |
- |
PREDICTED: EID1-like F-box protein 3 [Jatropha curcas] |
| 9 |
Hb_004109_150 |
0.2124572688 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 10 |
Hb_171900_030 |
0.2131300535 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein MYBAS2 [Jatropha curcas] |
| 11 |
Hb_010180_050 |
0.2133106935 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 12 |
Hb_006358_030 |
0.2144443915 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 13 |
Hb_003654_020 |
0.2152120674 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_005772_020 |
0.2153710474 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 15 |
Hb_003970_010 |
0.2196678591 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_000032_590 |
0.2207092461 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 17 |
Hb_002030_100 |
0.2242501082 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 18 |
Hb_000594_150 |
0.2248781759 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 19 |
Hb_012814_030 |
0.2273557415 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 20 |
Hb_002211_020 |
0.2281353079 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |