| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
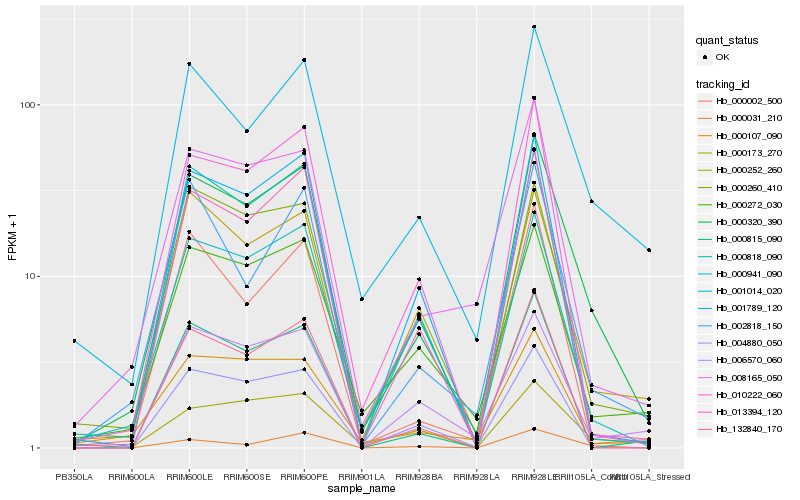
Hb_000320_390 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 2 |
Hb_001014_020 |
0.1152802979 |
- |
- |
gene GA family protein [Populus trichocarpa] |
| 3 |
Hb_013394_120 |
0.122846973 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 4 |
Hb_010222_060 |
0.1302940351 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000272_030 |
0.1337134152 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000173_270 |
0.1337216316 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000260_410 |
0.1340614634 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 8 |
Hb_000941_090 |
0.1347281188 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 9 |
Hb_008165_050 |
0.1349037436 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 10 |
Hb_001789_120 |
0.1352167501 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000818_090 |
0.1359534081 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 12 |
Hb_132840_170 |
0.1376411428 |
transcription factor |
TF Family: NAC |
Transcription factor [Theobroma cacao] |
| 13 |
Hb_000815_090 |
0.1432663302 |
- |
- |
PREDICTED: uncharacterized protein LOC105630464 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004880_050 |
0.143624063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002818_150 |
0.1441561539 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 16 |
Hb_006570_060 |
0.1447573976 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 17 |
Hb_000252_260 |
0.1458718397 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 18 |
Hb_000002_500 |
0.146133251 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000107_090 |
0.1481097067 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 20 |
Hb_000031_210 |
0.1488635184 |
- |
- |
cytochrome P450, putative [Ricinus communis] |