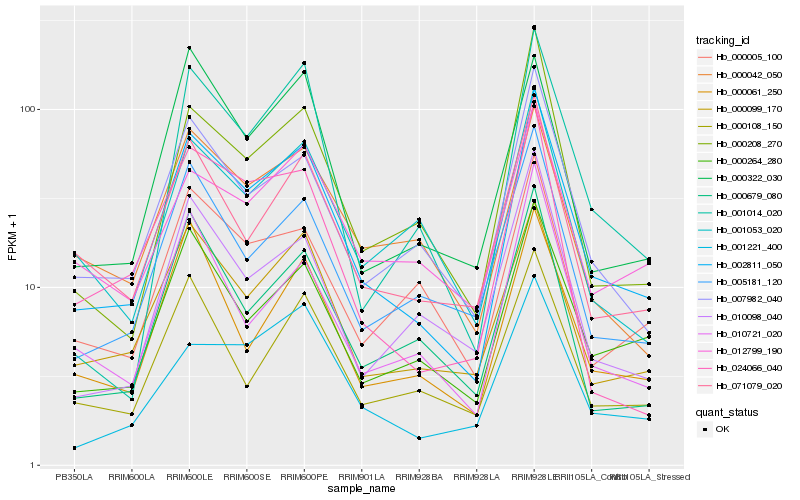
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 2 |
Hb_007982_040 |
0.1101350019 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000099_170 |
0.1110111925 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 4 |
Hb_000264_280 |
0.1158810025 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1-like [Jatropha curcas] |
| 5 |
Hb_071079_020 |
0.1204517232 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 6 |
Hb_000208_270 |
0.1237742383 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 7 |
Hb_005181_120 |
0.1243648245 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000061_250 |
0.1285339905 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 9 |
Hb_000679_080 |
0.1314315618 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001221_400 |
0.1328408271 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 11 |
Hb_000108_150 |
0.1336423466 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 12 |
Hb_000005_100 |
0.1353839663 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001014_020 |
0.1368728624 |
- |
- |
gene GA family protein [Populus trichocarpa] |
| 14 |
Hb_010721_020 |
0.1370410388 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_010098_040 |
0.1377336331 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 16 |
Hb_000042_050 |
0.1380819256 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001053_020 |
0.138107217 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 18 |
Hb_012799_190 |
0.1384182757 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 19 |
Hb_000322_030 |
0.1390051035 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 20 |
Hb_024066_040 |
0.1393592208 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |