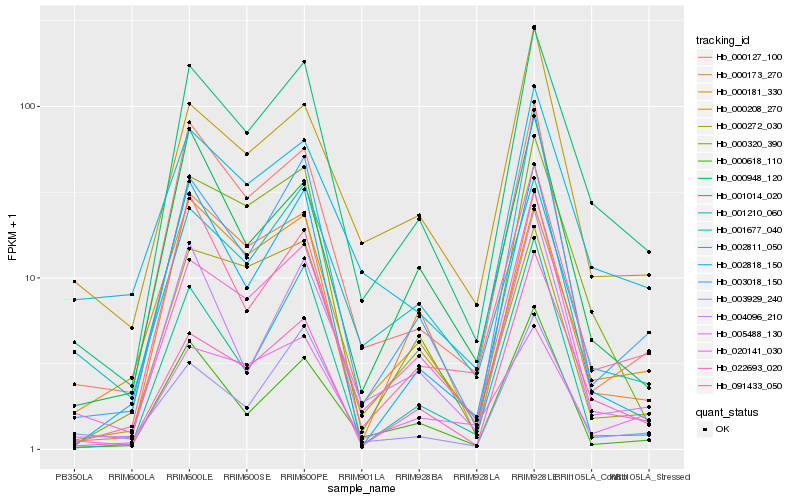
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001014_020 |
0.0 |
- |
- |
gene GA family protein [Populus trichocarpa] |
| 2 |
Hb_000320_390 |
0.1152802979 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 3 |
Hb_020141_030 |
0.1220113281 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_003018_150 |
0.1267239533 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 5 |
Hb_000127_100 |
0.1270298367 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 6 |
Hb_002811_050 |
0.1368728624 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 7 |
Hb_091433_050 |
0.1372874365 |
- |
- |
PREDICTED: uncharacterized protein LOC105646626 [Jatropha curcas] |
| 8 |
Hb_022693_020 |
0.1381213153 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 9 |
Hb_004096_210 |
0.1397836347 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001677_040 |
0.1421343291 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000181_330 |
0.1422127262 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 12 |
Hb_002818_150 |
0.1425993542 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 13 |
Hb_000272_030 |
0.1432832291 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000618_110 |
0.1440952556 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 15 |
Hb_000173_270 |
0.1447931325 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003929_240 |
0.1468656914 |
- |
- |
PREDICTED: uncharacterized protein LOC105643616 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000208_270 |
0.1482601959 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 18 |
Hb_000948_120 |
0.1484883696 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001210_060 |
0.1488142458 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 20 |
Hb_005488_130 |
0.149119902 |
- |
- |
transporter, putative [Ricinus communis] |