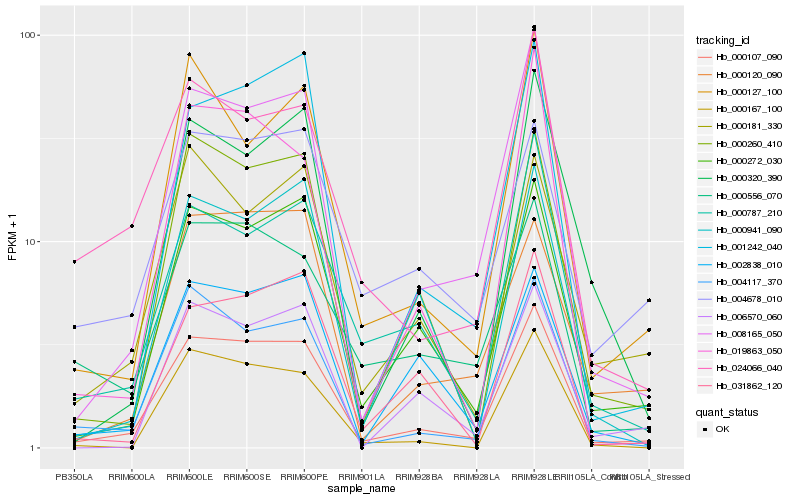
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_090 |
0.0 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 2 |
Hb_008165_050 |
0.0957092451 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_000272_030 |
0.1034978134 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004117_370 |
0.1125251896 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 5 |
Hb_000260_410 |
0.1250440872 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 6 |
Hb_000787_210 |
0.1380484944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_024066_040 |
0.1391839799 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 8 |
Hb_000127_100 |
0.1394392343 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 9 |
Hb_031862_120 |
0.1406875093 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 10 |
Hb_001242_040 |
0.1407462568 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 11 |
Hb_000167_100 |
0.1411775955 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_002838_010 |
0.1413958677 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 13 |
Hb_000181_330 |
0.1420656116 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 14 |
Hb_000120_090 |
0.1464138035 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 15 |
Hb_006570_060 |
0.1465960245 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 16 |
Hb_019863_050 |
0.1478458467 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 17 |
Hb_000320_390 |
0.1481097067 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 18 |
Hb_000556_070 |
0.1486399744 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At2g45590 [Jatropha curcas] |
| 19 |
Hb_000941_090 |
0.1503668748 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 20 |
Hb_004678_010 |
0.1542553298 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |