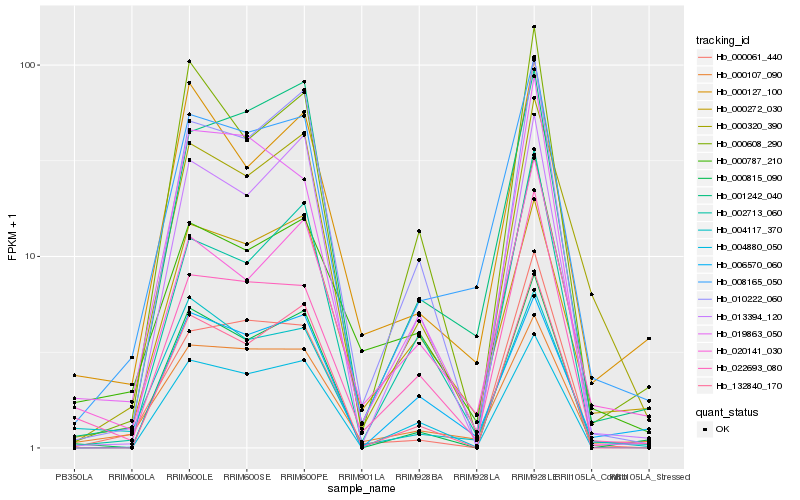
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008165_050 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_000107_090 |
0.0957092451 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 3 |
Hb_001242_040 |
0.127525574 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 4 |
Hb_000320_390 |
0.1349037436 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 5 |
Hb_000272_030 |
0.1356305637 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000127_100 |
0.1383651827 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 7 |
Hb_004117_370 |
0.1385653422 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 8 |
Hb_020141_030 |
0.1422761831 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 9 |
Hb_010222_060 |
0.1435726207 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000815_090 |
0.1441439703 |
- |
- |
PREDICTED: uncharacterized protein LOC105630464 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004880_050 |
0.1466409082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_019863_050 |
0.1469615613 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 13 |
Hb_006570_060 |
0.1475029841 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 14 |
Hb_022693_080 |
0.1476588564 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 15 |
Hb_002713_060 |
0.1487476272 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 16 |
Hb_132840_170 |
0.1488939897 |
transcription factor |
TF Family: NAC |
Transcription factor [Theobroma cacao] |
| 17 |
Hb_000061_440 |
0.1492330201 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_013394_120 |
0.1494605233 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 19 |
Hb_000608_290 |
0.1520069918 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 20 |
Hb_000787_210 |
0.1520278766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |