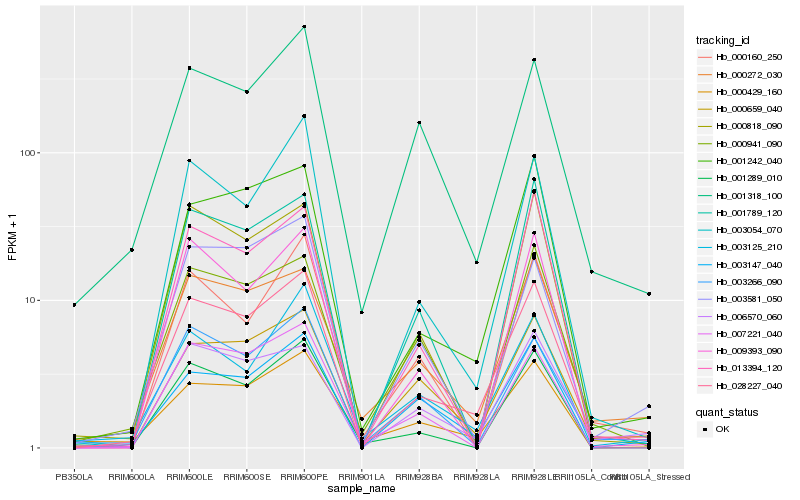
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028227_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 2 |
Hb_001242_040 |
0.1047811666 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 3 |
Hb_000272_030 |
0.1180426483 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000659_040 |
0.1184422963 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_006570_060 |
0.1207783064 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 6 |
Hb_003054_070 |
0.1221224075 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_013394_120 |
0.1222024032 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 8 |
Hb_009393_090 |
0.1241640149 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 9 |
Hb_003147_040 |
0.1262782288 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 10 |
Hb_000818_090 |
0.1264110786 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 11 |
Hb_003266_090 |
0.12753497 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 12 |
Hb_001289_010 |
0.1276209667 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Vitis vinifera] |
| 13 |
Hb_001789_120 |
0.1279847412 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000429_160 |
0.1295971577 |
- |
- |
PREDICTED: uncharacterized protein LOC105645177 [Jatropha curcas] |
| 15 |
Hb_000941_090 |
0.1334832289 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 16 |
Hb_000160_250 |
0.1352949591 |
- |
- |
Uncharacterized protein TCM_019235 [Theobroma cacao] |
| 17 |
Hb_007221_040 |
0.1381379841 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 isoform X2 [Populus euphratica] |
| 18 |
Hb_003581_050 |
0.13817881 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |
| 19 |
Hb_003125_210 |
0.1384248128 |
- |
- |
Phospholipase C 4 precursor, putative [Ricinus communis] |
| 20 |
Hb_001318_100 |
0.1400505152 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |