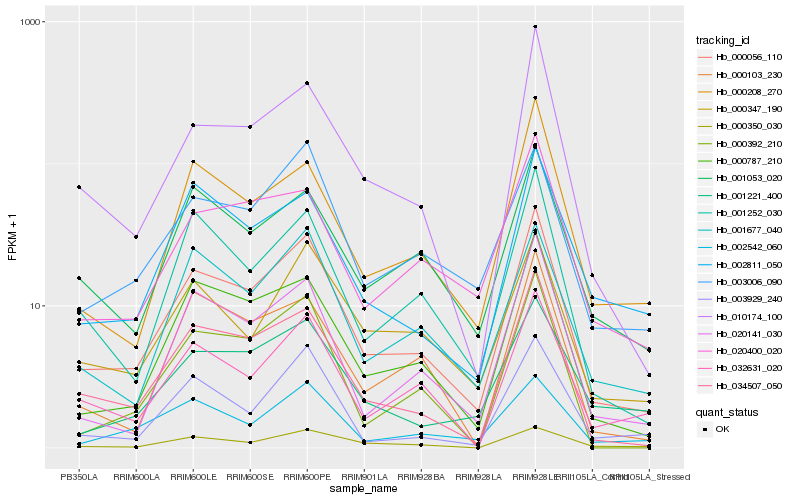
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000056_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 2 |
Hb_034507_050 |
0.0940616731 |
- |
- |
PREDICTED: uncharacterized protein LOC105646633 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000787_210 |
0.100778719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010174_100 |
0.1183300629 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 5 |
Hb_001252_030 |
0.1202396053 |
- |
- |
Serine/threonine-protein kinase Nek8, putative [Ricinus communis] |
| 6 |
Hb_020141_030 |
0.1208178072 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_001677_040 |
0.124000166 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 8 |
Hb_032631_020 |
0.1260543146 |
- |
- |
glycine-rich family protein [Populus trichocarpa] |
| 9 |
Hb_003006_090 |
0.1264619685 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 10 |
Hb_000208_270 |
0.1269159978 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 11 |
Hb_001053_020 |
0.1342392796 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 12 |
Hb_000347_190 |
0.1355877176 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 13 |
Hb_003929_240 |
0.1356553179 |
- |
- |
PREDICTED: uncharacterized protein LOC105643616 isoform X2 [Jatropha curcas] |
| 14 |
Hb_020400_020 |
0.1367401056 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 15 |
Hb_000392_210 |
0.1376102158 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0004s04600g [Populus trichocarpa] |
| 16 |
Hb_000350_030 |
0.1376840986 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 17 |
Hb_002811_050 |
0.1394475782 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 18 |
Hb_001221_400 |
0.1419155966 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 19 |
Hb_002542_060 |
0.1438873 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 20 |
Hb_000103_230 |
0.1439373427 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |