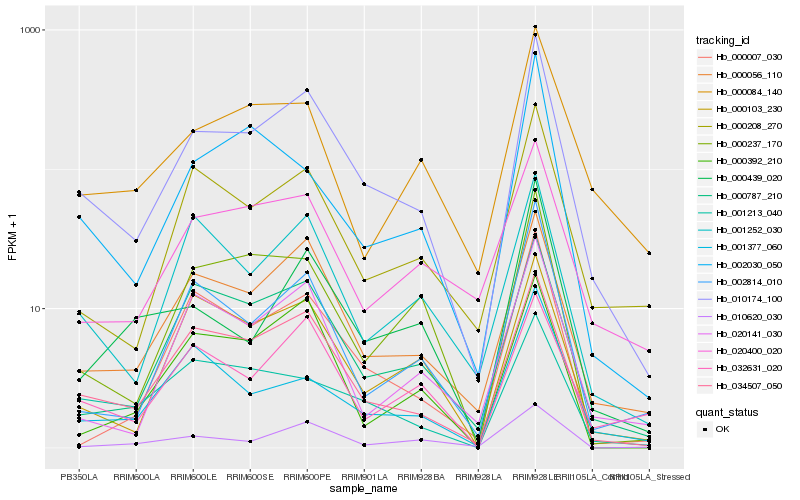
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010174_100 |
0.0 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 2 |
Hb_034507_050 |
0.0980902281 |
- |
- |
PREDICTED: uncharacterized protein LOC105646633 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000056_110 |
0.1183300629 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 4 |
Hb_000787_210 |
0.1428515729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000208_270 |
0.1529200654 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 6 |
Hb_002814_010 |
0.1584195279 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001252_030 |
0.1594927856 |
- |
- |
Serine/threonine-protein kinase Nek8, putative [Ricinus communis] |
| 8 |
Hb_000103_230 |
0.165497778 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000237_170 |
0.1659986787 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 10 |
Hb_000007_030 |
0.1672681415 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_020141_030 |
0.1672891829 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 12 |
Hb_020400_020 |
0.1683890464 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 13 |
Hb_032631_020 |
0.1720725277 |
- |
- |
glycine-rich family protein [Populus trichocarpa] |
| 14 |
Hb_000084_140 |
0.1723601751 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 15 |
Hb_002030_050 |
0.1749106902 |
- |
- |
classical arabinogalactan protein 7 precursor [Jatropha curcas] |
| 16 |
Hb_001213_040 |
0.17673004 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 17 |
Hb_001377_060 |
0.1770466057 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000439_020 |
0.177447383 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3-like [Jatropha curcas] |
| 19 |
Hb_010620_030 |
0.1781121281 |
- |
- |
beta-amyrin synthase [Euphorbia tirucalli] |
| 20 |
Hb_000392_210 |
0.1793431873 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0004s04600g [Populus trichocarpa] |