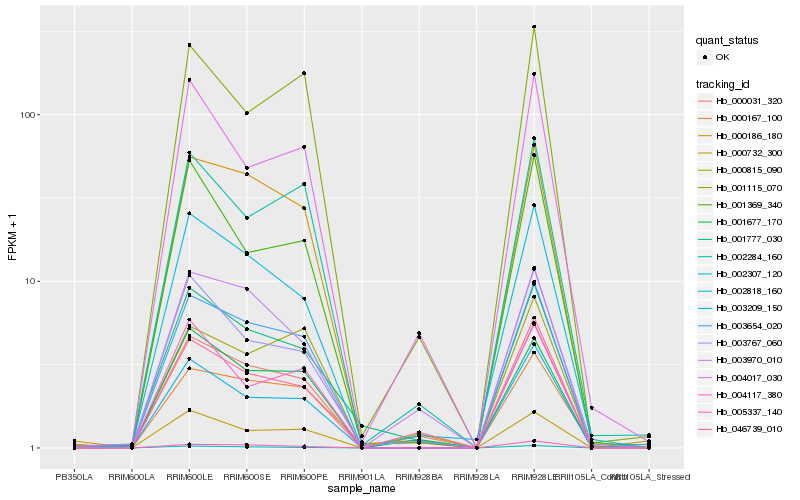
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003654_020 |
0.0 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000186_180 |
0.0549248152 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 3 |
Hb_003209_150 |
0.0840655055 |
- |
- |
CLE9, putative [Ricinus communis] |
| 4 |
Hb_000732_300 |
0.0915188985 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000031_320 |
0.0976447532 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002307_120 |
0.09919621 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 7 |
Hb_002818_160 |
0.0996228497 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 8 |
Hb_002284_160 |
0.105779614 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_001677_170 |
0.1073126087 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 10 |
Hb_005337_140 |
0.1100152484 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase 1-like [Jatropha curcas] |
| 11 |
Hb_001115_070 |
0.1126922127 |
- |
- |
RecName: Full=CASP-like protein 2C1; Short=PtCASPL2C1 [Populus trichocarpa] |
| 12 |
Hb_001777_030 |
0.1206908834 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 13 |
Hb_004117_380 |
0.1213966401 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 14 |
Hb_000167_100 |
0.1230094961 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_003767_060 |
0.125516168 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 16 |
Hb_046739_010 |
0.1281008948 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001369_340 |
0.1327481738 |
- |
- |
Peroxisomal membrane protein 11B [Theobroma cacao] |
| 18 |
Hb_004017_030 |
0.1328794521 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 19 |
Hb_000815_090 |
0.1361398337 |
- |
- |
PREDICTED: uncharacterized protein LOC105630464 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003970_010 |
0.136351747 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |