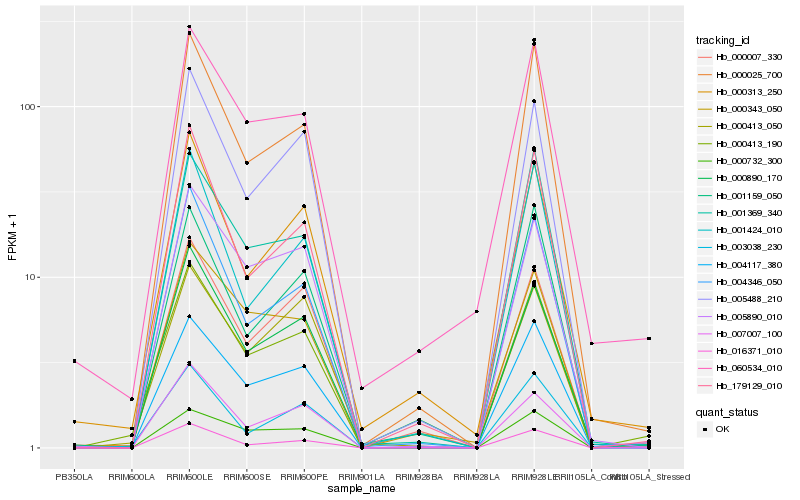
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000413_190 |
0.0 |
- |
- |
PREDICTED: oxygen-evolving enhancer protein 3-2, chloroplastic [Pyrus x bretschneideri] |
| 2 |
Hb_004117_380 |
0.0933837448 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 3 |
Hb_000025_700 |
0.0987585848 |
- |
- |
PREDICTED: serine--glyoxylate aminotransferase [Nelumbo nucifera] |
| 4 |
Hb_005488_210 |
0.1031148095 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 5 |
Hb_000732_300 |
0.1079491207 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004346_050 |
0.1102287425 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 7 |
Hb_007007_100 |
0.1102498748 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 8 |
Hb_179129_010 |
0.1118661182 |
- |
- |
hypothetical protein POPTR_0018s10470g [Populus trichocarpa] |
| 9 |
Hb_016371_010 |
0.1133751729 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 10 |
Hb_005890_010 |
0.1138705741 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 11 |
Hb_000007_330 |
0.1146824948 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 12 |
Hb_003038_230 |
0.1150207454 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 7 [Jatropha curcas] |
| 13 |
Hb_060534_010 |
0.1151433919 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 14 |
Hb_001369_340 |
0.115508607 |
- |
- |
Peroxisomal membrane protein 11B [Theobroma cacao] |
| 15 |
Hb_000413_050 |
0.1155855206 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000343_050 |
0.1159362539 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000313_250 |
0.1185516834 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 18 |
Hb_001159_050 |
0.1243391878 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 19 |
Hb_001424_010 |
0.1256003253 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000890_170 |
0.1264234558 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |