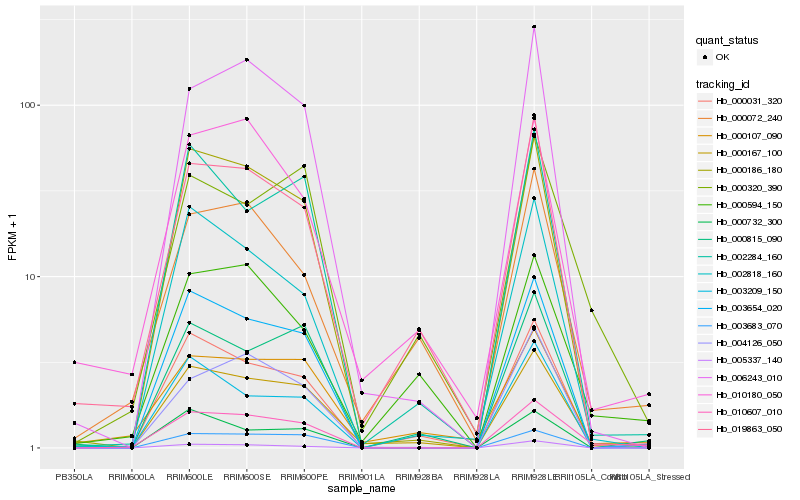
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010607_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 2 |
Hb_003654_020 |
0.1363996816 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000186_180 |
0.1415609641 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 4 |
Hb_000167_100 |
0.154447129 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_003683_070 |
0.1638288538 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 6 |
Hb_006243_010 |
0.1639554875 |
- |
- |
PREDICTED: cell wall protein IFF6 [Populus euphratica] |
| 7 |
Hb_000320_390 |
0.1676883559 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 8 |
Hb_002818_160 |
0.17092272 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 9 |
Hb_000031_320 |
0.1712238542 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_000732_300 |
0.1721712576 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005337_140 |
0.1724245104 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: wall-associated receptor kinase 1-like [Jatropha curcas] |
| 12 |
Hb_000107_090 |
0.1762503785 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 13 |
Hb_002284_160 |
0.1765048567 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_004126_050 |
0.1766236252 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VII.1 [Tarenaya hassleriana] |
| 15 |
Hb_000594_150 |
0.1775174277 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 16 |
Hb_000815_090 |
0.1799410758 |
- |
- |
PREDICTED: uncharacterized protein LOC105630464 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000072_240 |
0.1804238406 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_019863_050 |
0.1822396127 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 19 |
Hb_003209_150 |
0.1824514125 |
- |
- |
CLE9, putative [Ricinus communis] |
| 20 |
Hb_010180_050 |
0.1827633158 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |