| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
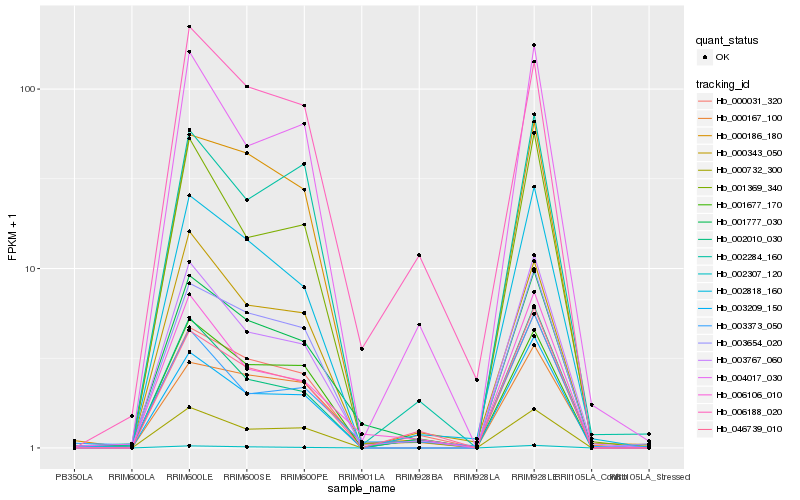
Hb_001777_030 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 2 |
Hb_003767_060 |
0.1011934433 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 3 |
Hb_002818_160 |
0.1029685326 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 4 |
Hb_000167_100 |
0.1057422972 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_006106_010 |
0.1061662902 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_000031_320 |
0.1080143889 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000186_180 |
0.1148356193 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 8 |
Hb_001677_170 |
0.1179422199 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 9 |
Hb_003654_020 |
0.1206908834 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_003209_150 |
0.1222074373 |
- |
- |
CLE9, putative [Ricinus communis] |
| 11 |
Hb_002307_120 |
0.1226569289 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 12 |
Hb_004017_030 |
0.1250504616 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 13 |
Hb_003373_050 |
0.1265054213 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 14 |
Hb_046739_010 |
0.1287482034 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006188_020 |
0.1302076413 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 16 |
Hb_002010_030 |
0.1318504516 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 17 |
Hb_001369_340 |
0.1324632663 |
- |
- |
Peroxisomal membrane protein 11B [Theobroma cacao] |
| 18 |
Hb_000343_050 |
0.1338614862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000732_300 |
0.1347445588 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002284_160 |
0.134990411 |
- |
- |
glyceraldehyde 3-phosphate dehydrogenase, putative [Ricinus communis] |