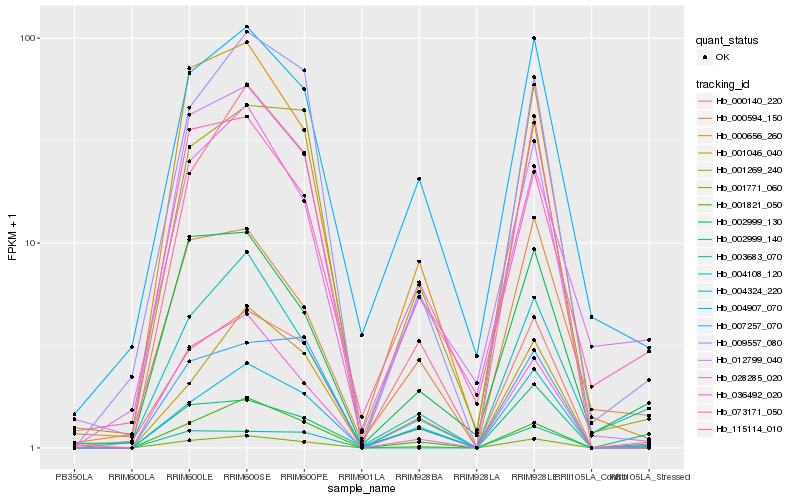
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001771_060 |
0.0 |
- |
- |
hypothetical protein B456_012G144100 [Gossypium raimondii] |
| 2 |
Hb_004108_120 |
0.1197619985 |
- |
- |
PREDICTED: EID1-like F-box protein 3 [Jatropha curcas] |
| 3 |
Hb_002999_130 |
0.141484189 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 4 |
Hb_028285_020 |
0.1521319637 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_002999_140 |
0.1553578982 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 6 |
Hb_009557_080 |
0.1558727863 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 7 |
Hb_000656_260 |
0.1597004549 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 8 |
Hb_073171_050 |
0.1613799914 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 9 |
Hb_115114_010 |
0.1615261167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000140_220 |
0.1645383979 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 11 |
Hb_001046_040 |
0.1647455335 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_004907_070 |
0.1650104035 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 13 |
Hb_004324_220 |
0.1661312322 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 14 |
Hb_001821_050 |
0.1677611735 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 15 |
Hb_001269_240 |
0.168311645 |
- |
- |
aquaporin NIP6.1 family protein [Populus trichocarpa] |
| 16 |
Hb_003683_070 |
0.1692199002 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 17 |
Hb_000594_150 |
0.1707481123 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 18 |
Hb_007257_070 |
0.1710749656 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 19 |
Hb_012799_040 |
0.1713518816 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 20 |
Hb_036492_020 |
0.1739601251 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |