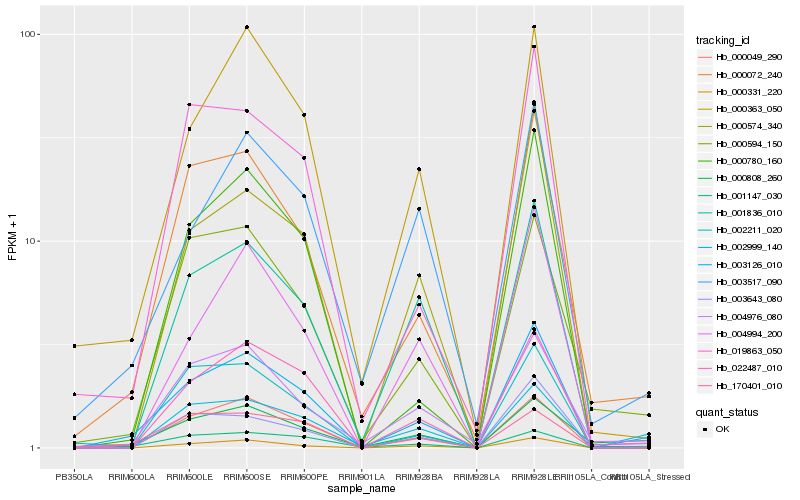
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003126_010 |
0.0 |
- |
- |
PREDICTED: oligopeptide transporter 7-like isoform X1 [Malus domestica] |
| 2 |
Hb_000072_240 |
0.1239620468 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000049_290 |
0.1490425797 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_022487_010 |
0.154562597 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 5 |
Hb_000331_220 |
0.1588991493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000363_050 |
0.1597142978 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000594_150 |
0.1600458671 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 8 |
Hb_000808_260 |
0.1603071543 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 9 |
Hb_000780_160 |
0.1609139571 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 10 |
Hb_019863_050 |
0.1619078405 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 11 |
Hb_004976_080 |
0.1635289415 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 12 |
Hb_003517_090 |
0.1675183802 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000574_340 |
0.1682898089 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 14 |
Hb_002999_140 |
0.1685889383 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 15 |
Hb_001836_010 |
0.1701924731 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 16 |
Hb_004994_200 |
0.1717281622 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 17 |
Hb_170401_010 |
0.1724675479 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 18 |
Hb_003643_080 |
0.173197159 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 19 |
Hb_001147_030 |
0.1737992219 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 20 |
Hb_002211_020 |
0.1739829552 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |