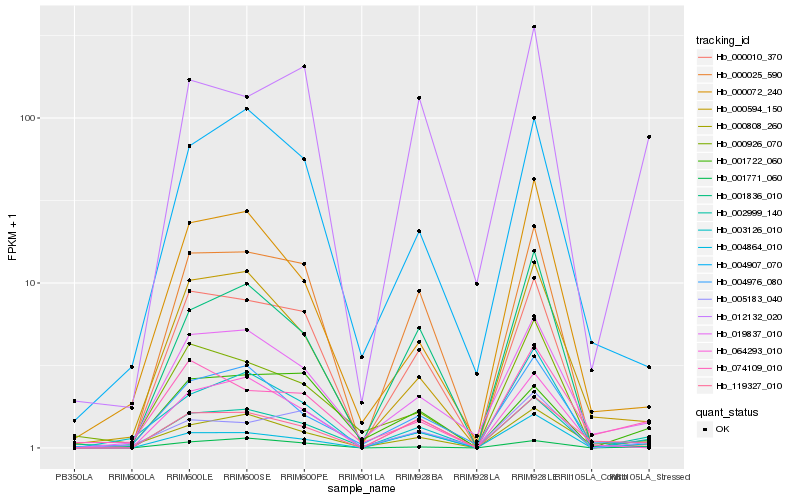
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002999_140 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 2 |
Hb_019837_010 |
0.137433213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001771_060 |
0.1553578982 |
- |
- |
hypothetical protein B456_012G144100 [Gossypium raimondii] |
| 4 |
Hb_000808_260 |
0.1621329058 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 5 |
Hb_004864_010 |
0.1643085833 |
- |
- |
hypothetical protein CICLE_v10003960mg, partial [Citrus clementina] |
| 6 |
Hb_003126_010 |
0.1685889383 |
- |
- |
PREDICTED: oligopeptide transporter 7-like isoform X1 [Malus domestica] |
| 7 |
Hb_000594_150 |
0.1700971628 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 8 |
Hb_000010_370 |
0.1701104962 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 9 |
Hb_012132_020 |
0.1704766616 |
- |
- |
glutamine synthetase [Hevea brasiliensis] |
| 10 |
Hb_004976_080 |
0.1714153145 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 11 |
Hb_000926_070 |
0.179951268 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_119327_010 |
0.1811897231 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 13 |
Hb_074109_010 |
0.1820468824 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 14 |
Hb_001722_060 |
0.183115777 |
- |
- |
- |
| 15 |
Hb_005183_040 |
0.1845197113 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 16 |
Hb_001836_010 |
0.1846035385 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 17 |
Hb_000025_590 |
0.1852306341 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000072_240 |
0.1853776562 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_004907_070 |
0.1877992838 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 20 |
Hb_064293_010 |
0.189562203 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |