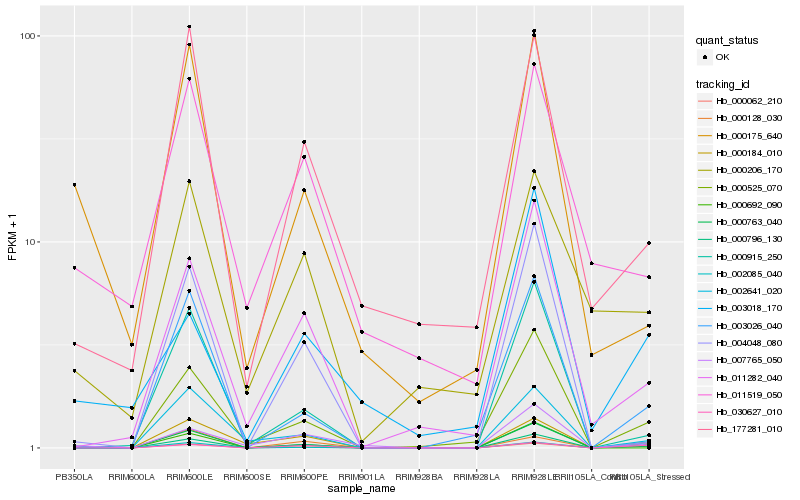
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000796_130 |
0.0 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 2 |
Hb_000062_210 |
0.1997742787 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Pyrus x bretschneideri] |
| 3 |
Hb_030627_010 |
0.2109566604 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 4 |
Hb_007765_050 |
0.2259974976 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000525_070 |
0.2277490686 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like [Jatropha curcas] |
| 6 |
Hb_002085_040 |
0.2382697855 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 7 |
Hb_000763_040 |
0.2409865019 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 8 |
Hb_000175_640 |
0.2443398612 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003026_040 |
0.2501908417 |
- |
- |
Uncharacterized protein TCM_007861 [Theobroma cacao] |
| 10 |
Hb_011282_040 |
0.2519433493 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 11 |
Hb_000128_030 |
0.2524666165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000184_010 |
0.2591771116 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 13 |
Hb_177281_010 |
0.2661693797 |
- |
- |
PREDICTED: 50S ribosomal protein 5, chloroplastic-like [Citrus sinensis] |
| 14 |
Hb_000692_090 |
0.2667819756 |
- |
- |
- |
| 15 |
Hb_004048_080 |
0.2677503251 |
- |
- |
hypothetical protein POPTR_0001s40360g [Populus trichocarpa] |
| 16 |
Hb_002641_020 |
0.268047983 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 17 |
Hb_003018_170 |
0.269488608 |
- |
- |
hypothetical protein POPTR_0005s11660g [Populus trichocarpa] |
| 18 |
Hb_000206_170 |
0.2696933463 |
- |
- |
hypothetical protein JCGZ_25514 [Jatropha curcas] |
| 19 |
Hb_000915_250 |
0.2737609681 |
- |
- |
chloroplast latex aldolase-like protein [Manihot esculenta] |
| 20 |
Hb_011519_050 |
0.2747501347 |
- |
- |
hypothetical protein POPTR_0015s04810g [Populus trichocarpa] |