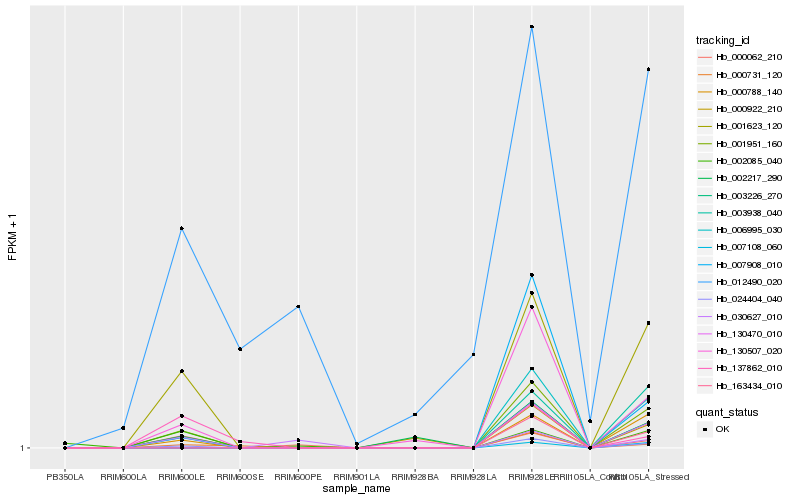
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001623_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001951_160 |
0.0905796203 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 3 |
Hb_024404_040 |
0.1221486116 |
- |
- |
hypothetical protein JCGZ_22351 [Jatropha curcas] |
| 4 |
Hb_000788_140 |
0.1270742387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_130507_020 |
0.2383155489 |
- |
- |
- |
| 6 |
Hb_000922_210 |
0.2576568975 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002217_290 |
0.2578485526 |
- |
- |
hypothetical protein JCGZ_19038 [Jatropha curcas] |
| 8 |
Hb_000062_210 |
0.2622680048 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Pyrus x bretschneideri] |
| 9 |
Hb_002085_040 |
0.2684651336 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 10 |
Hb_000731_120 |
0.2952075002 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 11 |
Hb_137862_010 |
0.3007988979 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_007908_010 |
0.3176613388 |
- |
- |
- |
| 13 |
Hb_012490_020 |
0.3231885857 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 14 |
Hb_007108_060 |
0.3264445395 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_006995_030 |
0.3273620494 |
- |
- |
PREDICTED: uncharacterized protein LOC102663993 [Glycine max] |
| 16 |
Hb_030627_010 |
0.3281092326 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |
| 17 |
Hb_003226_270 |
0.3314468066 |
- |
- |
hypothetical protein POPTR_0014s12390g [Populus trichocarpa] |
| 18 |
Hb_130470_010 |
0.3315376298 |
- |
- |
hypothetical protein, partial [Pseudomonas tolaasii] |
| 19 |
Hb_003938_040 |
0.3319908856 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 20 |
Hb_163434_010 |
0.3335151369 |
- |
- |
hypothetical protein JCGZ_13470 [Jatropha curcas] |