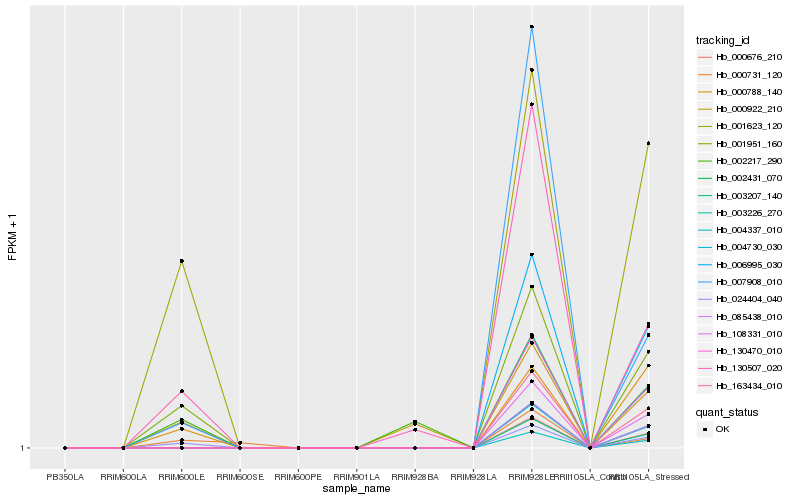
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_160 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 2 |
Hb_024404_040 |
0.0362720266 |
- |
- |
hypothetical protein JCGZ_22351 [Jatropha curcas] |
| 3 |
Hb_001623_120 |
0.0905796203 |
- |
- |
PREDICTED: uncharacterized protein LOC105650382 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000788_140 |
0.1219934665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_130507_020 |
0.1717673923 |
- |
- |
- |
| 6 |
Hb_007908_010 |
0.2325035694 |
- |
- |
- |
| 7 |
Hb_000922_210 |
0.2390082695 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002217_290 |
0.2400899635 |
- |
- |
hypothetical protein JCGZ_19038 [Jatropha curcas] |
| 9 |
Hb_000731_120 |
0.2553989011 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 10 |
Hb_006995_030 |
0.2639203749 |
- |
- |
PREDICTED: uncharacterized protein LOC102663993 [Glycine max] |
| 11 |
Hb_003226_270 |
0.2641141002 |
- |
- |
hypothetical protein POPTR_0014s12390g [Populus trichocarpa] |
| 12 |
Hb_130470_010 |
0.2641369665 |
- |
- |
hypothetical protein, partial [Pseudomonas tolaasii] |
| 13 |
Hb_163434_010 |
0.2647621173 |
- |
- |
hypothetical protein JCGZ_13470 [Jatropha curcas] |
| 14 |
Hb_108331_010 |
0.264988401 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 15 |
Hb_085438_010 |
0.2655069338 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 16 |
Hb_002431_070 |
0.265517155 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA11 [Jatropha curcas] |
| 17 |
Hb_004730_030 |
0.2655468296 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 18 |
Hb_000676_210 |
0.2659019587 |
- |
- |
Cytochrome P450 superfamily protein [Theobroma cacao] |
| 19 |
Hb_003207_140 |
0.2659379316 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 20 |
Hb_004337_010 |
0.2663078007 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |